Population structure and genetic connectivity of the scalloped hammerhead shark (Sphyrna lewini) across nursery grounds from the Eastern Tropical Pacific: Implications for management and conservation
- PMID: 36525407
- PMCID: PMC9757582
- DOI: 10.1371/journal.pone.0264879
Population structure and genetic connectivity of the scalloped hammerhead shark (Sphyrna lewini) across nursery grounds from the Eastern Tropical Pacific: Implications for management and conservation
Abstract
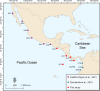
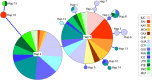
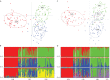
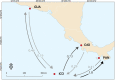
Defining demographically independent units and understanding patterns of gene flow between them is essential for managing and conserving exploited populations. The critically endangered scalloped hammerhead shark, Sphyrna lewini, is a coastal semi-oceanic species found worldwide in tropical and subtropical waters. Pregnant females give birth in shallow coastal estuarine habitats that serve as nursery grounds for neonates and small juveniles, whereas adults move offshore and become highly migratory. We evaluated the population structure and connectivity of S. lewini in coastal areas and one oceanic island (Cocos Island) across the Eastern Tropical Pacific (ETP) using both sequences of the mitochondrial DNA control region (mtCR) and 9 nuclear-encoded microsatellite loci. The mtCR defined two genetically discrete groups: one in the Mexican Pacific and another one in the central-southern Eastern Tropical Pacific (Guatemala, Costa Rica, Panama, and Colombia). Overall, the mtCR data showed low levels of haplotype diversity ranging from 0.000 to 0.608, while nucleotide diversity ranged from 0.000 to 0.0015. More fine-grade population structure was detected using microsatellite loci where Guatemala, Costa Rica, and Panama differed significantly. Relatedness analysis revealed that individuals within nursery areas were more closely related than expected by chance, suggesting that S. lewini may exhibit reproductive philopatric behaviour within the ETP. Findings of at least two different management units, and evidence of philopatric behaviour call for intensive conservation actions for this highly threatened species in the ETP.
Copyright: © 2022 Elizondo-Sancho et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


References
-
- Dulvy NK, Sadovy Y, Reynolds JD. Extinction vulnerability in marine populations. Fish Fish. 2003.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

