IL7 genetic variation and toxicity to immune checkpoint blockade in patients with melanoma
- PMID: 36526722
- PMCID: PMC9800275
- DOI: 10.1038/s41591-022-02095-5
IL7 genetic variation and toxicity to immune checkpoint blockade in patients with melanoma
Abstract
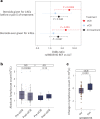
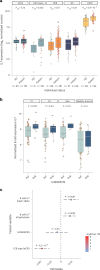
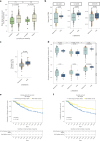
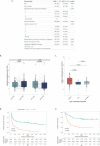
Treatment with immune checkpoint blockade (ICB) frequently triggers immune-related adverse events (irAEs), causing considerable morbidity. In 214 patients receiving ICB for melanoma, we observed increased severe irAE risk in minor allele carriers of rs16906115, intronic to IL7. We found that rs16906115 forms a B cell-specific expression quantitative trait locus (eQTL) to IL7 in patients. Patients carrying the risk allele demonstrate increased pre-treatment B cell IL7 expression, which independently associates with irAE risk, divergent immunoglobulin expression and more B cell receptor mutations. Consistent with the role of IL-7 in T cell development, risk allele carriers have distinct ICB-induced CD8+ T cell subset responses, skewing of T cell clonality and greater proportional repertoire occupancy by large clones. Finally, analysis of TCGA data suggests that risk allele carriers independently have improved melanoma survival. These observations highlight key roles for B cells and IL-7 in both ICB response and toxicity and clinical outcomes in melanoma.
© 2022. The Author(s).
Conflict of interest statement
C.A.T., R.A.W., O.T., W.Y., R.C., A.V.A., P.K.S., I.N., J.J.G. and A.G. declare no competing interests. M.R.M. reports grants from Roche, grants from AstraZeneca, grants and personal fees from GSK, personal fees and other from Novartis, other from Millenium, personal fees and other from Immunocore, personal fees and other from BMS, personal fees and other from Eisai, other from Pfizer, personal fees, non-financial support and other from Merck/MSD, personal fees and other from Rigontec (acquired by MSD), other from Regeneron, personal fees and other from BiolineRx, personal fees and other from Array Biopharma (now Pfizer), non-financial support and other from Replimune, personal fees from Kineta and personal fees from Silicon Therapeutics, all outside the submitted work. D.C. reports funding from Haliodx and consultancy for Merck. K.S. reports grants from Gilead, Eli Lilly and BMS and personal fees from UCB, AZD, Abbvie and Eli Lilly. P.E. performed consultancy for Abbvie, AstraZeneca, BMS, Boehringer Ingelheim, Galapagos, Gilead, Janssen, MSD, Eli Lilly, Novartis, Pfizer, Roche and Samsung and was involved in clinical trials for Abbvie, BMS, Eli Lilly, Novartis, Pfizer, Roche and Samsung. T.K.C. reports research/advisory boards/consultancy/honorarium (institutional and personal, paid and unpaid) from AstraZeneca, Aveo, Bayer, BMS, Eisai, EMD Serono, Exelixis, GSK, IQVA, Ipsen, Kanaph, Eli Lilly, Merck, Nikang, Novartis, Pfizer, Roche, Sanofi/Aventis, Takeda and Tempest. Travel, accommodations, expenses, medical writing in relation to consulting, advisory roles or honoraria. Stock options: Pionyr and Tempest. Other: up-to-date royalties, CME-related events (for example, OncLIve, PVI and MJH Life Sciences) honoraria. NCI GU Steering Committee. Patents filed, royalties or other intellectual properties (no income as of current date): related to biomarkers of immune checkpoint blockers and ctDNA. No speaker’s bureau. B.P.F. received conference support from BMS and performed consultancy for UCB.
Figures





Comment in
-
Using genetics to predict toxicity of cancer immunotherapy.Nat Med. 2022 Dec;28(12):2471-2472. doi: 10.1038/s41591-022-02096-4. Nat Med. 2022. PMID: 36526721 No abstract available.
References
-
- Larkin J, et al. Five-year survival with combined nivolumab and ipilimumab in advanced melanoma. N. Engl. J. Med. 2019;381:1535–1546. - PubMed
-
- Weber J, et al. Adjuvant nivolumab versus ipilimumab in resected stage III or IV melanoma. N. Engl. J. Med. 2017;377:1824–1835. - PubMed
-
- Robert C, et al. Pembrolizumab versus ipilimumab in advanced melanoma (KEYNOTE-006): post-hoc 5-year results from an open-label, multicentre, randomised, controlled, phase 3 study. Lancet Oncol. 2019;20:1239–1251. - PubMed
-
- Michot JM, et al. Immune-related adverse events with immune checkpoint blockade: a comprehensive review. Eur. J. Cancer. 2016;54:139–148. - PubMed
-
- Schneider BJ, et al. Management of immune-related adverse events in patients treated with immune checkpoint inhibitor therapy: ASCO guideline update. J. Clin. Oncol. 2021;39:4073–4126. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

