Mutations in SARS-CoV-2 structural proteins: a global analysis
- PMID: 36528612
- PMCID: PMC9759450
- DOI: 10.1186/s12985-022-01951-7
Mutations in SARS-CoV-2 structural proteins: a global analysis
Abstract
Background: Emergence of new variants mainly variants of concerns (VOC) is caused by mutations in main structural proteins of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). Therefore, we aimed to investigate the mutations among structural proteins of SARS-CoV-2 globally.
Methods: We analyzed samples of amino-acid sequences (AASs) for envelope (E), membrane (M), nucleocapsid (N), and spike (S) proteins from the declaration of the coronavirus 2019 (COVID-19) as pandemic to January 2022. The presence and location of mutations were then investigated by aligning the sequences to the reference sequence and categorizing them based on frequency and continent. Finally, the related human genes with the viral structural genes were discovered, and their interactions were reported.
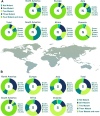
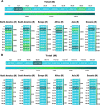
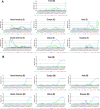
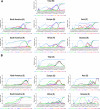
Results: The results indicated that the most relative mutations among the E, M, N, and S AASs occurred in the regions of 7 to 14, 66 to 88, 164 to 205, and 508 to 635 AAs, respectively. The most frequent mutations in E, M, N, and S proteins were T9I, I82T, R203M/R203K, and D614G. D614G was the most frequent mutation in all six geographical areas. Following D614G, L18F, A222V, E484K, and N501Y, respectively, were ranked as the most frequent mutations in S protein globally. Besides, A-kinase Anchoring Protein 8 Like (AKAP8L) was shown as the linkage unit between M, E, and E cluster genes.
Conclusion: Screening the structural protein mutations can help scientists introduce better drug and vaccine development strategies.
Keywords: COVID-19; Evolutionary analysis; Genome-wide mutations; Mutations; SARS-CoV-2.
© 2022. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures








References
-
- World Health Organization. https://www.who.int/news-room/detail/27-04-2020-who-timeline---covid-19, 2020.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

