Molecular evolution of the ependymin-related gene epdl2 in African weakly electric fish
- PMID: 36529459
- PMCID: PMC9997568
- DOI: 10.1093/g3journal/jkac331
Molecular evolution of the ependymin-related gene epdl2 in African weakly electric fish
Abstract
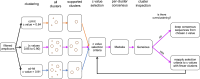
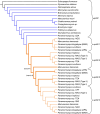
Gene duplication and subsequent molecular evolution can give rise to taxon-specific gene specializations. In previous work, we found evidence that African weakly electric fish (Mormyridae) may have as many as three copies of the epdl2 gene, and the expression of two epdl2 genes is correlated with electric signal divergence. Epdl2 belongs to the ependymin-related family (EPDR), a functionally diverse family of secretory glycoproteins. In this study, we first describe vertebrate EPDR evolution and then present a detailed evolutionary history of epdl2 in Mormyridae with emphasis on the speciose genus Paramormyrops. Using Sanger sequencing, we confirm three apparently functional epdl2 genes in Paramormyrops kingsleyae. Next, we developed a nanopore-based amplicon sequencing strategy and bioinformatics pipeline to obtain and classify full-length epdl2 gene sequences (N = 34) across Mormyridae. Our phylogenetic analysis proposes three or four epdl2 paralogs dating from early Paramormyrops evolution. Finally, we conducted selection tests which detected positive selection around the duplication events and identified ten sites likely targeted by selection in the resulting paralogs. These sites' locations in our modeled 3D protein structure involve four sites in ligand binding and six sites in homodimer formation. Together, these findings strongly imply an evolutionary mechanism whereby epdl2 genes underwent selection-driven functional specialization after tandem duplications in the rapidly speciating Paramormyrops. Considering previous evidence, we propose that epdl2 may contribute to electric signal diversification in mormyrids, an important aspect of species recognition during mating.
Keywords: electric fish; ependymin; gene duplication; molecular evolution.
© The Author(s) 2022. Published by Oxford University Press on behalf of the Genetics Society of America.
Conflict of interest statement
Conflicts of interest None declared.
Figures





References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
