Chromosome-scale genome assembly of Eustoma grandiflorum, the first complete genome sequence in the genus Eustoma
- PMID: 36529465
- PMCID: PMC9911058
- DOI: 10.1093/g3journal/jkac329
Chromosome-scale genome assembly of Eustoma grandiflorum, the first complete genome sequence in the genus Eustoma
Abstract
Eustoma grandiflorum (Raf.) Shinn. is an annual herbaceous plant native to the southern United States, Mexico, and the Greater Antilles. It has a large flower with a variety of colors and is an important flower crop. In this study, we established a chromosome-scale de novo assembly of E. grandiflorum genome sequences by integrating four genomic and genetic approaches: (1) Pacific Biosciences (PacBio) Sequel deep sequencing, (2) error correction of the assembly by Illumina short reads, (3) scaffolding by chromatin conformation capture sequencing (Hi-C), and (4) genetic linkage maps derived from an F2 mapping population. Thirty-six pseudomolecules and 64 unplaced scaffolds were created, with a total length of 1,324.8 Mb. A total of 36,619 genes were predicted on the genome as high-confidence genes. A comparison of genome structure between E. grandiflorum and C. canephora or O. pumila suggested whole-genome duplication after the divergence between the families Gentianaceae and Rubiaceae. Phylogenetic analysis with single-copy genes suggested that the divergence time between Gentianaceae and Rubiaceae was 74.94 MYA. Genetic diversity analysis was performed for nine commercial E. grandiflorum varieties bred in Japan, from which 254,205 variants were identified. This first report on the construction of a reference genome sequence in the genus Eustoma is expected to contribute to genetic and genomic studies in this genus and in the family Gentianaceae.
Keywords: Eustoma grandiflorum; gene prediction; genetic diversity; genome assembly.
© The Author(s) 2022. Published by Oxford University Press on behalf of the Genetics Society of America.
Conflict of interest statement
Conflicts of interest The authors declare no conflicts of interest associated with this manuscript.
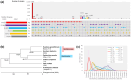
Figures

References
-
- Barba-Gonzalez R, Tapia-Campos E, Lara-Bañuelos TY, Cepeda-Cornejo V, Dupre P, Arratia-Ramirez G. Interspecific hybridization advances in the genus Eustoma. In International Society for Horticultural Science (ISHS), editor. VIII International Symposium on New Ornamental Crops and XII International Protea Research Symposium. Belgium: Leuven; 2015. p. 93–100.
-
- Bickhart DM, Rosen BD, Koren S, Sayre BL, Hastie AR, Chan S, Lee J, Lam ET, Liachko I, Sullivan ST, et al. . Single-molecule sequencing and chromatin conformation capture enable de novo reference assembly of the domestic goat genome. Nat Genet. 2017;49(4):643–650. doi:10.1038/ng.3802. - DOI - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
