A Polymeric Nanoparticle Formulation for Targeted mRNA Delivery to Fibroblasts
- PMID: 36529964
- PMCID: PMC9929262
- DOI: 10.1002/advs.202205475
A Polymeric Nanoparticle Formulation for Targeted mRNA Delivery to Fibroblasts
Abstract
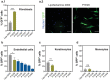
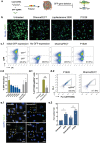
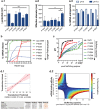
Messenger RNA (mRNA)-based therapies offer enhanced control over the production of therapeutic proteins for many diseases. Their clinical implementation warrants formulations capable of delivering them safely and effectively to target sites. Owing to their chemical versatility, polymeric nanoparticles can be designed by combinatorial synthesis of different ionizable, cationic, and aromatic moieties to modulate cell targeting, using inexpensive formulation steps. Herein, 152 formulations are evaluated by high-throughput screening using a reporter fibroblast model sensitive to functional delivery of mRNA encoding Cre recombinase. Using in vitro and in vivo models, a polymeric nanoformulation based on the combination of 3 specific monomers is identified to transfect fibroblasts much more effectively than other cell types populating the skin, with superior performance than lipid-based transfection agents in the delivery of Cas9 mRNA and guide RNA. This tropism can be explained by receptor-mediated endocytosis, involving CD26 and FAP, which are overexpressed in profibrotic fibroblasts. Structure-activity analysis reveals that efficient mRNA delivery required the combination of high buffering capacity and low mRNA binding affinity for rapid release upon endosomal escape. These results highlight the use of high-throughput screening to rapidly identify chemical features towards the design of highly efficient mRNA delivery systems targeting fibrotic diseases.
Keywords: CRISPR/Cas9; gene edition; high-throughput screening; messenger RNA; polymeric nanoparticles.
© 2022 The Authors. Advanced Science published by Wiley-VCH GmbH.
Conflict of interest statement
The authors declare the following competing interests: AFR and LF are applicants of a provisional Portuguese patent no. 117455. The remaining authors declare no conflict of interest.
Figures



References
-
- Ginn S. L., Amaya A. K., Alexander I. E., Edelstein M., Abedi M. R., J. Gene Med. 2018, 20, e3015. - PubMed
-
- Yin H., Kanasty R. L., Eltoukhy A. A., Vegas A. J., Dorkin J. R., Anderson D. G., Nat. Rev. Genet. 2014, 15, 541. - PubMed
-
- Akinc A., Maier M. A., Manoharan M., Fitzgerald K., Jayaraman M., Barros S., Ansell S., Du X., Hope M. J., Madden T. D., Mui B. L., Semple S. C., Tam Y. K., Ciufolini M., Witzigmann D., Kulkarni J. A., Meel v. d. R., Cullis P. R., Nat. Nanotechnol. 2019, 14, 1084. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous
