Critical Review of Plant-Derived Compounds as Possible Inhibitors of SARS-CoV-2 Proteases: A Comparison with Experimentally Validated Molecules
- PMID: 36530229
- PMCID: PMC9753184
- DOI: 10.1021/acsomega.2c05766
Critical Review of Plant-Derived Compounds as Possible Inhibitors of SARS-CoV-2 Proteases: A Comparison with Experimentally Validated Molecules
Abstract
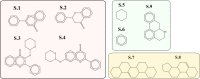
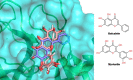
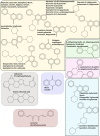
Ever since coronavirus disease 2019 (COVID-19), caused by SARS-CoV-2, was declared a pandemic on March 11, 2020, by the WHO, a concerted effort has been made to find compounds capable of acting on the virus and preventing its replication. In this context, researchers have refocused part of their attention on certain natural compounds that have shown promising effects on the virus. Considering the importance of this topic in the current context, this study aimed to present a critical review and analysis of the main reports of plant-derived compounds as possible inhibitors of the two SARS-CoV-2 proteases: main protease (Mpro) and Papain-like protease (PLpro). From the search in the PubMed database, a total of 165 published articles were found that met the search patterns. A total of 590 unique molecules were identified from a total of 122 articles as potential protease inhibitors. At the same time, 114 molecules reported as natural products and with annotation of theoretical support and antiviral effects were extracted from the COVID-19 Help database. After combining the molecules extracted from articles and those obtained from the database, we identified 648 unique molecules predicted as potential inhibitors of Mpro and/or PLpro. According to our results, several of the predicted compounds with higher theoretical confidence are present in many plants used in traditional medicine and even food, such as flavonoids, carboxylic acids, phenolic acids, triterpenes, terpenes phytosterols, and triterpenoids. These are potential inhibitors of Mpro and PLpro. Although the predictions of several molecules against SARS-CoV-2 are promising, little experimental information was found regarding certain families of compounds. Only 45 out of the 648 unique molecules have experimental data validating them as inhibitors of Mpro or PLpro, with the most frequent scaffold present in these 45 compounds being the flavone. The novelty of this work lies in the analysis of the structural diversity of the chemical space among the molecules predicted as inhibitors of SARS-CoV-2 Mpro and PLpro proteases and the comparison to those molecules experimentally validated. This work emphasizes the need for experimental validation of certain families of compounds, preferentially combining classical enzymatic assays with interaction-based methods. Furthermore, we recommend checking the presence of Pan-Assay Interference Compounds (PAINS) and the presence of molecules previously reported as inhibitors of Mpro or PLpro to optimize resources and time in the discovery of new SARS-CoV-2 antivirals from plant-derived molecules.
© 2022 The Authors. Published by American Chemical Society.
Conflict of interest statement
The authors declare no competing financial interest.
Figures
References
-
- WHO . WHO Timeline - COVID-19; https://www.who.int/news/item/27-04-2020-who-timeline—covid-19 (accessed August 25, 2022).
-
- Jin Z.; Du X.; Xu Y.; Deng Y.; Liu M.; Zhao Y.; Zhang B.; Li X.; Zhang L.; Peng C.; Duan Y.; Yu J.; Wang L.; Yang K.; Liu F.; Jiang R.; Yang X.; You T.; Liu X.; Yang X.; Bai F.; Liu H.; Liu X.; Guddat L. W.; Xu W.; Xiao G.; Qin C.; Shi Z.; Jiang H.; Rao Z.; Yang H. Structure of Mpro from SARS-CoV-2 and Discovery of Its Inhibitors. Nature 2020, 582 (7811), 289–293. 10.1038/s41586-020-2223-y. - DOI - PubMed
-
- Harcourt B. H.; Jukneliene D.; Kanjanahaluethai A.; Bechill J.; Severson K. M.; Smith C. M.; Rota P. A.; Baker S. C. Identification of Severe Acute Respiratory Syndrome Coronavirus Replicase Products and Characterization of Papain-Like Protease Activity. J. Virol 2004, 78 (24), 13600–13612. 10.1128/JVI.78.24.13600-13612.2004. - DOI - PMC - PubMed
-
- Frieman M.; Ratia K.; Johnston R. E.; Mesecar A. D.; Baric R. S. Severe Acute Respiratory Syndrome Coronavirus Papain-Like Protease Ubiquitin-Like Domain and Catalytic Domain Regulate Antagonism of IRF3 and NF-KB Signaling. J. Virol 2009, 83 (13), 6689–6705. 10.1128/JVI.02220-08. - DOI - PMC - PubMed
Publication types
LinkOut - more resources
Full Text Sources
Miscellaneous

