A novel panel of clinically relevant miRNAs signature accurately differentiates oral cancer from normal mucosa
- PMID: 36531016
- PMCID: PMC9753689
- DOI: 10.3389/fonc.2022.1072579
A novel panel of clinically relevant miRNAs signature accurately differentiates oral cancer from normal mucosa
Abstract
Introduction: Although a considerable body of knowledge has been accumulated regarding the early diagnosis and treatment of oral squamous cell carcinoma (OSCC), its survival rates have not improved over the last decades. Thus, deciphering the molecular mechanisms governing oral cancer will support the development of even better diagnostic and therapeutic strategies. Previous studies have linked aberrantly expressed microRNAs (miRNAs) with the development of OSCC.
Methods: We combined bioinformatical and molecular methods to identify miRNAs with possible clinical significance as biomarkers in OSCC. A set of 10 miRNAs were selected via an in silico approach by analysing the 3'untranslated regions (3'UTRs) of cancer-related mRNAs such as FLRT2, NTRK3, and SLC8A1, TFCP2L1 and etc. RT-qPCR was used to compare the expression of in silico identified miRNAs in OSCC and normal tissues (n=32).
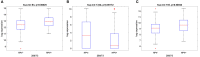
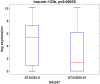
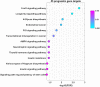
Results: Among the screened miRNAs, miR-21-5p (p < 0.0001), miR-93-5p (p < 0.0197), miR-146b-5p (p <0.0012), miR-155-5p (p < 0.0001), miR-182-5p (p < 0.0001) were significantly overexpressed, whereas miR-133b (p < 0.05) was significantly downregulated in OSCC tissues, a scenario confirmed in two additional OSCC validation cohorts: Regina Elena National Cancer Institute (IRE cohort, N=74) and The Cancer Genome Atlas Data Portal (TCGA cohort, N=354). Initial stage tumors (T1, T2) expressed significantly higher levels of miR-133b (p < 0.0004) compared to more advanced ones (T3, T4). Also, we identified miR-93-5p (p < 0.0003), miR-133b (p < 0.0017) and miR-155-5p (p < 0.0004) as correlated with HPV-induced OSCC. The high expression of these 6 miRNAs as a signature predicted shorter disease-free survival (DFS) and could efficiently distinguish OSCC cases from healthy controls with areas under the curve (AUC) of 0.91 with sensitivity and specificity of 0.98 and 0.6, respectively. Further target identification analysis revealed enrichment of genes involved in FOXO, longevity, glycan biosynthesis and p53 cancer-related signaling pathways. Also, the selected targets were underexpressed in OSCC tissues and showed clinical significance related to overall survival (OS) and DFS.
Discussion: Our results demonstrate that a novel panel consisting of miR-21-5p, miR-93-5p, miR-133b, miR-146b-5p, miR-155-5p and miR-182-5p could be used as OSCC-specific molecular signature with diagnostic and prognostic significance related to OS and DFS.
Keywords: biomarker; disease-free survival; mRNA; miRNA; oral squamous cell carcinoma; overall survival.
Copyright © 2022 Mehterov, Sacconi, Pulito, Vladimirov, Haralanov, Pazardjikliev, Nonchev, Berindan-Neagoe, Blandino and Sarafian.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures




References
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

