Two alternative splicing variants of a wheat gene TaNAK1, TaNAK1.1 and TaNAK1.2, differentially regulate flowering time and plant architecture leading to differences in seed yield of transgenic Arabidopsis
- PMID: 36531344
- PMCID: PMC9751850
- DOI: 10.3389/fpls.2022.1014176
Two alternative splicing variants of a wheat gene TaNAK1, TaNAK1.1 and TaNAK1.2, differentially regulate flowering time and plant architecture leading to differences in seed yield of transgenic Arabidopsis
Abstract
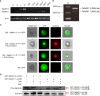
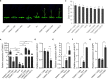
In wheat production, appropriate flowering time and ideal plant architecture are the prerequisites for high grain yield. Alternative splicing (AS) is a vital process that regulates gene expression at the post-transcriptional level, and AS events in wheat have been found to be closely related to grain-related traits and abiotic stress tolerance. However, AS events and their biological roles in regulating flowering time and plant architecture in wheat remain unclear. In this study, we report that TaNAK1 undergoes AS, producing three splicing variants. Molecular characterization of TaNAK1 and its splicing variants demonstrated that all three protein isoforms have a conserved NB-ARC domain and a protein kinase domain, but the positions of these two domains and the length of the protein kinase domains are different among them, implying that they may have different three-dimensional structures and therefore have different functions. Further investigations showed that the two splicing variants of TaNAK1, TaNAK1.1 and TaNAK1.2, exhibited different expression patterns during wheat growth and development, while the other one, TaNAK1.3, was not detected. Subcellular localization demonstrated that TaNAK1.1 was mainly localized in the cytoplasm, while TaNAK1.2 was localized in the nucleus and cytoplasm. Both TaNAK1.1 and TaNAK1.2 exhibit protein kinase activity in vitro. Ectopic expression of TaNAK1.1 and TaNAK1.2 in Arabidopsis demonstrated that these two splicing variants play opposite roles in regulating flowering time and plant architecture, resulting in different seed yields. TaNAK1.2 positive regulates the transition from vegetative to reproductive growth, plant height, branching number, seed size, and seed yield of Arabidopsis, while TaNAK1.1 negatively regulates these traits. Our findings provide new gene resource for regulating flowering time and plant architecture in crop breeding for high grain yield.
Keywords: TaNAK1; alternative splicing; flowering time; plant architecture; seed yield; wheat.
Copyright © 2022 Wu, Zhang, Hu, Zheng, Zhang, Liu, Ma and Zhao.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest. The reviewer HM declared a shared affiliation with the authors to the handling editor at the time of review.
Figures


References
-
- Ananda G. K. S., Norton S. L., Blomstedt C., Furtado A., Møller B. L., Gleadow R., et al. (2022). Transcript profiles of wild and domesticated sorghum under water-stressed conditions and the differential impact on dhurrin metabolism. Planta 255, 1–19. doi: 10.1007/s00425-022-03831-4 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Research Materials

