Gene expression analysis in endometriosis: Immunopathology insights, transcription factors and therapeutic targets
- PMID: 36532015
- PMCID: PMC9748153
- DOI: 10.3389/fimmu.2022.1037504
Gene expression analysis in endometriosis: Immunopathology insights, transcription factors and therapeutic targets
Abstract
Background: Endometriosis is recognized as an estrogen-dependent inflammation disorder, estimated to affect 8%-15% of women of childbearing age. Currently, the etiology and pathogenesis of endometriosis are not completely clear. Underlying mechanism for endometriosis is still under debate and needs further exploration. The involvement of transcription factors and immune mediations may be involved in the pathophysiological process of endometriosis, but the specific mechanism remains to be explored. This study aims to investigate the underlying molecular mechanisms in endometriosis.
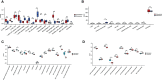
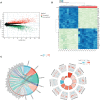
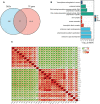
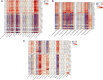
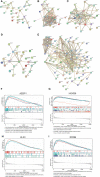
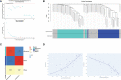
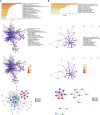
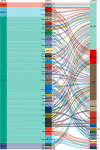
Methods: The gene expression profile of endometriosis was obtained from the gene expression omnibus (GEO) database. Gene set variation analysis (GSVA) and gene set enrichment analysis (GSEA) were applied to the endometriosis GSE7305 datasets. Cibersort and MCP-counter were used to explore the immune response gene sets, immune response pathway, and immune environment. Differentially expressed genes (DEGs) were identified and screened. Common biological pathways were being investigated using the kyoto encyclopedia of genes and genomes (KEGG) pathway enrichment analysis. Transcription factors were from The Human Transcription Factors. The least absolute shrinkage and selection operator (Lasso) model identified four differential expressions of transcription factors (AEBP1, HOXB6, KLF2, and RORB). Their diagnostic value was calculated by receiver operating characteristic (ROC) curve analysis and validated in the validation cohort (GSE11691, GSE23339). By constructing the interaction network of crucial transcription factors, weighted gene coexpression network analysis (WGCNA) was used to search for key module genes. Metascape was used for enrichment analysis of essential module genes and obtained HOXB6, KLF2. The HOXB6 and KLF2 were further verified as the only two intersection genes according to Support Vector Machine Recursive Feature Elimination (SVM-RFE) and random forest models. We constructed ceRNA (lncRNA-miRNA-mRNA) networks with four potential transcription factors. Finally, we performed molecular docking for goserelin and dienogest with four transcription factors (AEBP1, HOXB6, KLF2, and RORB) to screen potential drug targets.
Results: Immune and metabolic pathways were enriched in GSVA and GSEA. In single sample gene set enrichment analysis (ssGSEA), most immune infiltrating cells, immune response gene sets, and immune response pathways are differentially expressed between endometriosis and non-endometriosis. Twenty-seven transcription factors were screened from differentially expressed genes. Most of the twenty-seven transcription factors were correlated with immune infiltrating cells, immune response gene sets and immune response pathways. Furthermore, Adipocyte enhancer binding protein 1 (AEBP1), Homeobox B6 (HOXB6), Kruppel Like Factor 2 (KLF2) and RAR Related Orphan Receptor B (RORB) were selected out from twenty-seven transcription factors. ROC analysis showed that the four genes had a high diagnostic value for endometriosis. In addition, KLF2 and HOXB6 were found to play particularly important roles in multiple modules (String, WGCNA, SVM-RFE, random forest) on the gene interaction network. Using the ceRNA network, we found that NEAT1 may regulate the expressions of AEBP1, HOXB6 and RORB, while X Inactive Specific Transcript (XIST) may control the expressions of HOXB6, RORB and KLF2. Finally, we found that goserelin and dienogest may be potential drugs to regulate AEBP1, HOXB6, KLF2 and RORB through molecular docking.
Conclusions: AEBP1, HOXB6, KLF2, and RORB may be potential biomarkers for endometriosis. Two of them, KLF2 and HOXB6, are critical molecules in the gene interaction network of endometriosis. Discovered by molecular docking, AEBP1, HOXB6, KLF2, and RORB are targets for goserelin and dienogest.
Keywords: ceRNA network; endometriosis; goserelin and dienogest; immune disorder; transcription factors.
Copyright © 2022 Geng, Huang, Li, Guo, Wang, Zheng and Guo.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures




References
-
- Halme J, Hammond MG, Hulka JF, Raj SG, Talbert LM. Retrograde menstruation in healthy women and in patients with endometriosis. Obstet Gynecol (1984) 64:151–4. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

