Viral infectious diseases severity: co-presence of transcriptionally active microbes (TAMs) can play an integral role for disease severity
- PMID: 36532032
- PMCID: PMC9755851
- DOI: 10.3389/fimmu.2022.1056036
Viral infectious diseases severity: co-presence of transcriptionally active microbes (TAMs) can play an integral role for disease severity
Abstract
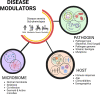
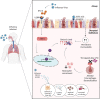
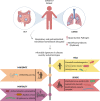
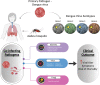
Humans have been challenged by infectious diseases for all of their recorded history, and are continually being affected even today. Next-generation sequencing (NGS) has enabled identification of, i) culture independent microbes, ii) emerging disease-causing pathogens, and iii) understanding of the genome architecture. This, in turn, has highlighted that pathogen/s are not a monolith, and thereby allowing for the differentiation of the wide-ranging disease symptoms, albeit infected by a primary pathogen. The conventional 'one disease - one pathogen' paradigm has been positively revisited by considering limited yet important evidence of the co-presence of multiple transcriptionally active microbes (TAMs), potential pathogens, in various infectious diseases, including the COVID-19 pandemic. The ubiquitous microbiota presence inside humans gives reason to hypothesize that the microbiome, especially TAMs, contributes to disease etiology. Herein, we discuss current evidence and inferences on the co-infecting microbes particularly in the diseases caused by the RNA viruses - Influenza, Dengue, and the SARS-CoV-2. We have highlighted that the specific alterations in the microbial taxonomic abundances (dysbiosis) is functionally connected to the exposure of primary infecting pathogen/s. The microbial presence is intertwined with the differential host immune response modulating differential disease trajectories. The microbiota-host interactions have been shown to modulate the host immune responses to Influenza and SARS-CoV-2 infection, wherein the active commensal microbes are involved in the generation of virus-specific CD4 and CD8 T-cells following the influenza virus infection. Furthermore, COVID-19 dysbiosis causes an increase in inflammatory cytokines such as IL-6, TNF-α, and IL-1β, which might be one of the important predisposing factors for severe infection. Through this article, we aim to provide a comprehensive view of functional microbiomes that can have a significant regulatory impact on predicting disease severity (mild, moderate and severe), as well as clinical outcome (survival and mortality). This can offer fresh perspectives on the novel microbial biomarkers for stratifying patients for severe disease symptoms, disease prevention and augmenting treatment regimens.
Keywords: RNA viruses; disease severity; immune response; infectious diseases; microbial biomarkers; microbiome; transcriptionally active microbes (TAMs).
Copyright © 2022 Yadav and Pandey.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

Similar articles
-
Dual RNA-Seq reveals transcriptionally active microbes (TAMs) dynamics in the serum of dengue patients associated with disease severity.Front Microbiol. 2023 Nov 30;14:1307859. doi: 10.3389/fmicb.2023.1307859. eCollection 2023. Front Microbiol. 2023. PMID: 38107870 Free PMC article.
-
Increased Abundance of Achromobacter xylosoxidans and Bacillus cereus in Upper Airway Transcriptionally Active Microbiome of COVID-19 Mortality Patients Indicates Role of Co-Infections in Disease Severity and Outcome.Microbiol Spectr. 2022 Jun 29;10(3):e0231121. doi: 10.1128/spectrum.02311-21. Epub 2022 May 17. Microbiol Spectr. 2022. PMID: 35579429 Free PMC article.
-
SARS-CoV-2 infection is associated with intestinal permeability, systemic inflammation, and microbial dysbiosis in hospitalized patients.Microbiol Spectr. 2024 Nov 5;12(11):e0068024. doi: 10.1128/spectrum.00680-24. Epub 2024 Sep 30. Microbiol Spectr. 2024. PMID: 39345212 Free PMC article.
-
Microbial co-infections in COVID-19: Associated microbiota and underlying mechanisms of pathogenesis.Microb Pathog. 2021 Jul;156:104941. doi: 10.1016/j.micpath.2021.104941. Epub 2021 May 4. Microb Pathog. 2021. PMID: 33962007 Free PMC article. Review.
-
Autoimmunity and COVID-19 - The microbiotal connection.Autoimmun Rev. 2021 Aug;20(8):102865. doi: 10.1016/j.autrev.2021.102865. Epub 2021 Jun 10. Autoimmun Rev. 2021. PMID: 34118455 Free PMC article. Review.
Cited by
-
Single-cell RNA-Seq reveals intracellular microbial diversity within immune cells during SARS-CoV-2 infection and recovery.iScience. 2023 Oct 30;26(11):108357. doi: 10.1016/j.isci.2023.108357. eCollection 2023 Nov 17. iScience. 2023. PMID: 38026191 Free PMC article.
-
Dual RNA-Seq reveals transcriptionally active microbes (TAMs) dynamics in the serum of dengue patients associated with disease severity.Front Microbiol. 2023 Nov 30;14:1307859. doi: 10.3389/fmicb.2023.1307859. eCollection 2023. Front Microbiol. 2023. PMID: 38107870 Free PMC article.
-
Analyses of Extended-Spectrum-β-Lactamase, Metallo-β-Lactamase, and AmpC-β-Lactamase Producing Enterobacteriaceae from the Dairy Value Chain in India.Antibiotics (Basel). 2023 Sep 14;12(9):1449. doi: 10.3390/antibiotics12091449. Antibiotics (Basel). 2023. PMID: 37760745 Free PMC article.
-
Recombinant serralysin metalloproteases D enhances the intracellular replication of infectious bovine rhinotracheitis virus.Front Microbiol. 2025 May 9;16:1567288. doi: 10.3389/fmicb.2025.1567288. eCollection 2025. Front Microbiol. 2025. PMID: 40415927 Free PMC article.
-
Single cell genomics based insights into the impact of cell-type specific microbial internalization on disease severity.Front Immunol. 2024 May 21;15:1401320. doi: 10.3389/fimmu.2024.1401320. eCollection 2024. Front Immunol. 2024. PMID: 38835769 Free PMC article. Review.
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

