Immune landscape-based machine-learning-assisted subclassification, prognosis, and immunotherapy prediction for glioblastoma
- PMID: 36532035
- PMCID: PMC9751405
- DOI: 10.3389/fimmu.2022.1027631
Immune landscape-based machine-learning-assisted subclassification, prognosis, and immunotherapy prediction for glioblastoma
Abstract
Introduction: As a malignant brain tumor, glioblastoma (GBM) is characterized by intratumor heterogeneity, a worse prognosis, and highly invasive, lethal, and refractory natures. Immunotherapy has been becoming a promising strategy to treat diverse cancers. It has been known that there are highly heterogeneous immunosuppressive microenvironments among different GBM molecular subtypes that mainly include classical (CL), mesenchymal (MES), and proneural (PN), respectively. Therefore, an in-depth understanding of immune landscapes among them is essential for identifying novel immune markers of GBM.
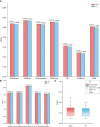
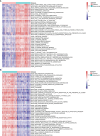
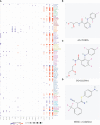
Methods and results: In the present study, based on collecting the largest number of 109 immune signatures, we aim to achieve a precise diagnosis, prognosis, and immunotherapy prediction for GBM by performing a comprehensive immunogenomic analysis. Firstly, machine-learning (ML) methods were proposed to evaluate the diagnostic values of these immune signatures, and the optimal classifier was constructed for accurate recognition of three GBM subtypes with robust and promising performance. The prognostic values of these signatures were then confirmed, and a risk score was established to divide all GBM patients into high-, medium-, and low-risk groups with a high predictive accuracy for overall survival (OS). Therefore, complete differential analysis across GBM subtypes was performed in terms of the immune characteristics along with clinicopathological and molecular features, which indicates that MES shows much higher immune heterogeneity compared to CL and PN but has significantly better immunotherapy responses, although MES patients may have an immunosuppressive microenvironment and be more proinflammatory and invasive. Finally, the MES subtype is proved to be more sensitive to 17-AAG, docetaxel, and erlotinib using drug sensitivity analysis and three compounds of AS-703026, PD-0325901, and MEK1-2-inhibitor might be potential therapeutic agents.
Conclusion: Overall, the findings of this research could help enhance our understanding of the tumor immune microenvironment and provide new insights for improving the prognosis and immunotherapy of GBM patients.
Keywords: glioblastoma (GBM); immune landscape; immunotherapy; machine-learning (ML); prognosis; subclassification.
Copyright © 2022 Li, He, Li, Li, Pu and Guo.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest. The handling editor LZ declared a shared parent affiliation with the authors HL, JH, ML, KL, XP, YG at the time of the review
Figures



Similar articles
-
Machine learning revealed stemness features and a novel stemness-based classification with appealing implications in discriminating the prognosis, immunotherapy and temozolomide responses of 906 glioblastoma patients.Brief Bioinform. 2021 Sep 2;22(5):bbab032. doi: 10.1093/bib/bbab032. Brief Bioinform. 2021. PMID: 33839757 Free PMC article.
-
Comprehensive Analysis of the Tumor Immune Microenvironment Landscape in Glioblastoma Reveals Tumor Heterogeneity and Implications for Prognosis and Immunotherapy.Front Immunol. 2022 Mar 2;13:820673. doi: 10.3389/fimmu.2022.820673. eCollection 2022. Front Immunol. 2022. PMID: 35309323 Free PMC article.
-
Genomics and proteomics to determine novel molecular subtypes and predict the response to immunotherapy and the effect of bevacizumab in glioblastoma.Sci Rep. 2024 Jul 31;14(1):17630. doi: 10.1038/s41598-024-68648-5. Sci Rep. 2024. PMID: 39085480 Free PMC article.
-
The PTEN-associated immune prognostic signature reveals the landscape of the tumor microenvironment in glioblastoma.J Neuroimmunol. 2023 Mar 15;376:578034. doi: 10.1016/j.jneuroim.2023.578034. Epub 2023 Jan 24. J Neuroimmunol. 2023. PMID: 36791582 Review.
-
Current Insights into Mesenchymal Signatures in Glioblastoma.Acta Med Okayama. 2022 Oct;76(5):489-502. doi: 10.18926/AMO/64024. Acta Med Okayama. 2022. PMID: 36352795 Review.
Cited by
-
Immune intrinsic escape signature stratifies prognosis, characterizes the tumor immune microenvironment, and identifies tumorigenic PPP1R8 in glioblastoma multiforme patients.Front Immunol. 2025 Aug 6;16:1577920. doi: 10.3389/fimmu.2025.1577920. eCollection 2025. Front Immunol. 2025. PMID: 40842996 Free PMC article.
-
Comprehensive machine learning-based integration develops a novel prognostic model for glioblastoma.Mol Ther Oncol. 2024 Jun 17;32(3):200838. doi: 10.1016/j.omton.2024.200838. eCollection 2024 Sep 19. Mol Ther Oncol. 2024. PMID: 39072291 Free PMC article.
-
Comprehensive understanding of glioblastoma molecular phenotypes: classification, characteristics, and transition.Cancer Biol Med. 2024 May 6;21(5):363-81. doi: 10.20892/j.issn.2095-3941.2023.0510. Cancer Biol Med. 2024. PMID: 38712813 Free PMC article. Review.
-
Lactylation-related gene signatures identify glioma molecular subtypes with prognostic, immunological, and therapeutic implications.Front Oncol. 2025 Jul 16;15:1613423. doi: 10.3389/fonc.2025.1613423. eCollection 2025. Front Oncol. 2025. PMID: 40740857 Free PMC article.
-
Artificial Intelligence-Assisted Drug and Biomarker Discovery for Glioblastoma: A Scoping Review of the Literature.Cancers (Basel). 2025 Feb 7;17(4):571. doi: 10.3390/cancers17040571. Cancers (Basel). 2025. PMID: 40002166 Free PMC article. Review.
References
-
- Verhaak RGW, Hoadley KA, Purdom E, Wang V, Qi Y, Wilkerson MD, et al. . Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in pdgfra, Idh1, egfr, and Nf1. Cancer Cell (2010) 17(1):98–110. doi: 10.1016/j.ccr.2009.12.020 - DOI - PMC - PubMed
-
- Soomro SH, Ting LR, Qing YY, Ren M. Molecular biology of glioblastoma: Classification and mutational locations. J Pak Med Assoc (2017) 67(9):1410–4. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

