STXBP3 and GOT2 predict immunological activity in acute allograft rejection
- PMID: 36532048
- PMCID: PMC9751189
- DOI: 10.3389/fimmu.2022.1025681
STXBP3 and GOT2 predict immunological activity in acute allograft rejection
Abstract
Background: Acute allograft rejection (AR) following renal transplantation contributes to chronic rejection and allograft dysfunction. The current diagnosis of AR remains dependent on renal allograft biopsy which cannot immediately detect renal allograft injury in the presence of AR. In this study, sensitive biomarkers for AR diagnosis were investigated and developed to protect renal function.
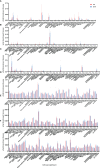
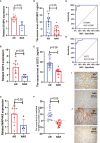
Methods: We analyzed pre- and postoperative data from five databases combined with our own data to identify the key differently expressed genes (DEGs). Furthermore, we performed a bioinformatics analysis to determine the immune characteristics of DEGs. The expression of key DEGs was further confirmed using the real-time quantitative PCR (RT-qPCR), enzyme-linked immunosorbent assay (ELISA), and immunohistochemical (IHC) staining in patients with AR. ROC curves analysis was used to estimate the performance of key DEGs in the early diagnosis of AR.
Results: We identified glutamic-oxaloacetic transaminase 2 (GOT2) and syntaxin binding protein 3 (STXBP3) as key DEGs. The higher expression of STXBP3 and GOT2 in patients with AR was confirmed using RT-qPCR, ELISA, and IHC staining. ROC curve analysis also showed favorable values of STXBP3 and GOT2 for the diagnosis of early stage AR.
Conclusions: STXBP3 and GOT2 could reflect the immunological status of patients with AR and have strong potential for the diagnosis of early-stage AR.
Keywords: GOT2; STXBP3; acute rejection; expression; kidney transplantation.
Copyright © 2022 Yao, Wang, Wang, Xiang, Chen, Zhou, Chen, Jiang and Chen.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



Similar articles
-
[Noninvasive diagnostic and predictive value in renal transplant recipients with acute rejection by measurement of urine Fractalkine].Zhonghua Yi Xue Za Zhi. 2017 Jan 10;97(2):92-98. doi: 10.3760/cma.j.issn.0376-2491.2017.02.003. Zhonghua Yi Xue Za Zhi. 2017. PMID: 28088951 Chinese.
-
Neutrophil gelatinase-associated lipocalin is a sensitive biomarker for the early diagnosis of acute rejection after living-donor kidney transplantation.Int Urol Nephrol. 2013 Aug;45(4):1159-67. doi: 10.1007/s11255-012-0321-y. Epub 2012 Nov 17. Int Urol Nephrol. 2013. PMID: 23161375
-
The identification of novel potential injury mechanisms and candidate biomarkers in renal allograft rejection by quantitative proteomics.Mol Cell Proteomics. 2014 Feb;13(2):621-31. doi: 10.1074/mcp.M113.030577. Epub 2013 Dec 12. Mol Cell Proteomics. 2014. PMID: 24335474 Free PMC article.
-
Currently available useful immunohistochemical markers of renal pathology for the diagnosis of renal allograft rejection.Nephrology (Carlton). 2015 Jul;20 Suppl 2:9-15. doi: 10.1111/nep.12460. Nephrology (Carlton). 2015. PMID: 26031579 Review.
-
Urine biomarkers informative of human kidney allograft rejection and tolerance.Hum Immunol. 2018 May;79(5):343-355. doi: 10.1016/j.humimm.2018.01.006. Epub 2018 Jan 31. Hum Immunol. 2018. PMID: 29366869 Review.
Cited by
-
Sex differences in brain protein expression and disease.Nat Med. 2023 Sep;29(9):2224-2232. doi: 10.1038/s41591-023-02509-y. Epub 2023 Aug 31. Nat Med. 2023. PMID: 37653343 Free PMC article.
-
GOT2: a moonlighting enzyme at the crossroads of cancer metabolism and theranostics.Front Immunol. 2025 Aug 6;16:1626914. doi: 10.3389/fimmu.2025.1626914. eCollection 2025. Front Immunol. 2025. PMID: 40842990 Free PMC article. Review.
-
Increasing Eligibility to Transplant Through the Selective Cytopheretic Device: A Review of Case Reports Across Multiple Clinical Conditions.Transplant Direct. 2024 May 16;10(6):e1627. doi: 10.1097/TXD.0000000000001627. eCollection 2024 Jun. Transplant Direct. 2024. PMID: 38769980 Free PMC article.
References
-
- Wolfe RA, Ashby VB, Milford EL, Ojo AO, Ettenger RE, Agodoa LY, et al. . Comparison of mortality in all patients on dialysis, patients on dialysis awaiting transplantation, and recipients of a first cadaveric transplant. N Engl J Med (1999) 341:1725–30. doi: 10.1056/nejm199912023412303 - DOI - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

