Comparative analysis of intestinal flora between rare wild red-crowned crane and white-naped crane
- PMID: 36532425
- PMCID: PMC9752901
- DOI: 10.3389/fmicb.2022.1007884
Comparative analysis of intestinal flora between rare wild red-crowned crane and white-naped crane
Abstract
Introduction: Animal intestines are extremely rich in microbial ecosystems. Numerous studies in different fields, such as epidemiology and histology, have revealed that gut microorganisms considerably mediate the survival and reproduction of animals. However, gut microbiology studies of homogeneously distributed wild cranes are still rare. This study aimed to understand the structural composition of the gut microbial community of wild cranes and elucidate the potential roles of the microorganisms.
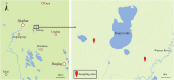
Methods: We used high-throughput sequencing to analyze the gut microbial community structure of wild cranes in the Zhalong Nature Reserve.
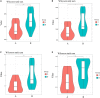
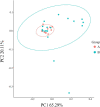
Results: A total of 1,965,683 valid tags and 5248 OTUs were obtained from 32 fecal samples. Twenty-six bacteria phyla and 523 genera were annotated from the intestinal tract of the red-crowned crane. Twenty-five bacteria phyla and 625 genera were annotated from the intestine of the white-naped crane. Firmicutes, Proteobacteria, and Bacteroidetes are the dominant bacterial phyla in the intestinal tract of red-crowned cranes, while Catellicoccus, Lactobacillus, Neisseria, and Streptococcus were the dominant genera. The dominant bacterial phyla in the intestinal tract of white-naped cranes were Firmicutes, Proteobacteria, Bacteroidetes, Epsilonbacteraeota, Actinobacteria, and Fusobacteria. However, the dominant genera were Catellicoccus, Lactobacillus, Neisseria, Campylobacter, Streptococcus, Anaerobiospirillum, Romboutsia, Turicibacter, Haemophilus, and Lautropia. Firmicutes had significantly higher relative abundance in the intestine of the red-crowned than white-naped cranes (P < 0.05). However, the relative abundance of Actinobacteria and Bacteroidetes was significantly higher (P < 0.05) in the intestines of white-naped than red-crowned cranes. The diversity of the intestinal flora between the two crane species was significantly different (P < 0.05). Besides, the alpha diversity of the intestinal flora was higher for white-naped than red-crowned cranes. Eight of the 41 functional pathways differed in the gut of both crane species (P < 0.05).
Discussion: Both species live in the same area and have similar feeding and behavioral characteristics. Therefore, host differences are possibly the main factors influencing the structural and functional differences in the composition of the gut microbial community. This study provides important reference data for constructing a crane gut microbial assessment system. The findings have implications for studying deeper relationships between crane gut microbes and genetics, nutrition, immunity, and disease.
Keywords: Zhalong Nature Reserve; high-throughput sequencing technology; intestinal microorganisms; red-crowned crane; white-naped crane.
Copyright © 2022 Gao, Song, Dong, Ji, Lei, Tian, Wu and Zou.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


References
-
- Byappanahalli M. N., Nevers M. B., Whitman R. L., Ge Z., Shively D., Spoljaric A., et al. (2015). Wildlife, urban inputs, and landscape configuration are responsible for degraded swimming water quality at an embayed beach. J. Great Lakes Res. 41 156–163. 10.1016/j.jglr.2014.11.027 - DOI
LinkOut - more resources
Full Text Sources

