Characterization of virulence and antimicrobial resistance genes of Aeromonas media strain SD/21-15 from marine sediments in comparison with other Aeromonas spp
- PMID: 36532448
- PMCID: PMC9752117
- DOI: 10.3389/fmicb.2022.1022639
Characterization of virulence and antimicrobial resistance genes of Aeromonas media strain SD/21-15 from marine sediments in comparison with other Aeromonas spp
Abstract
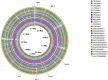
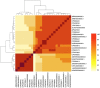
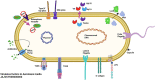
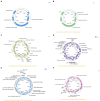
Aeromonas media is a Gram-negative bacterium ubiquitously found in aquatic environments. It is a foodborne pathogen associated with diarrhea in humans and skin ulceration in fish. In this study, we used whole genome sequencing to profile all antimicrobial resistance (AMR) and virulence genes found in A. media strain SD/21-15 isolated from marine sediments in Denmark. To gain a better understanding of virulence and AMR genes found in several A. media strains, we included 24 whole genomes retrieved from the public databanks whose isolates originate from different host species and environmental samples from Asia, Europe, and North America. We also compared the virulence genes of strain SD/21-15 with A. hydrophila, A. veronii, and A. salmonicida reference strains. We detected Msh pili, tap IV pili, and lateral flagella genes responsible for expression of motility and adherence proteins in all isolates. We also found hylA, hylIII, and TSH hemolysin genes in all isolates responsible for virulence in all isolates while the aerA gene was not detected in all A. media isolates but was present in A. hydrophila, A. veronii, and A. salmonicida reference strains. In addition, we detected LuxS and mshA-Q responsible for quorum sensing and biofilm formation as well as the ferric uptake regulator (Fur), heme and siderophore genes responsible for iron acquisition in all A. media isolates. As for the secretory systems, we found all genes that form the T2SS in all isolates while only the vgrG1, vrgG3, hcp, and ats genes that form parts of the T6SS were detected in some isolates. Presence of bla MOX-9 and bla OXA-427 β-lactamases as well as crp and mcr genes in all isolates is suggestive that these genes were intrinsically encoded in the genomes of all A. media isolates. Finally, the presence of various transposases, integrases, recombinases, virulence, and AMR genes in the plasmids examined in this study is suggestive that A. media has the potential to transfer virulence and AMR genes to other bacteria. Overall, we anticipate these data will pave way for further studies on virulence mechanisms and the role of A. media in the spread of AMR genes.
Keywords: Aeromonas media; antimicrobial resistance; intrinsic–extrinsic; plasmid; virulence; whole genome sequencing.
Copyright © 2022 Dubey, Ager-Wick, Peng, Evensen, Sørum and Munang’andu.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
References
-
- Abdel-Fattah G. M., Hafez E. E., Zaki M. E., Darwesh N. M. (2017). Cloning and expression of alpha hemolysin toxin gene of Staphylococcus aureus against human cancer tissue. Int. J. Appl. Sci. Biotechnol. 5, 22–29. doi: 10.3126/ijasbt.v5i1.17000 - DOI
-
- Alcock B. P., Raphenya A. R., Lau T. T., Tsang K. K., Bouchard M., Edalatmand A., et al. (2020). Nguyen A-LV, Cheng AA, Liu S: CARD 2020: antibiotic resistome surveillance with the comprehensive antibiotic resistance database. Nucleic Acids Res. 48, D517–D525. doi: 10.1093/nar/gkz935, PMID: - DOI - PMC - PubMed
-
- Allen D., Austin B., Colwell R. (1983). Aeromonas media, a new species isolated from river water. Int. J. Syst. Evol. Microbiol. 33, 599–604.
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

