Glutamine potentiates gentamicin to kill lab-evolved gentamicin-resistant and clinically isolated multidrug-resistant Escherichia coli
- PMID: 36532472
- PMCID: PMC9755591
- DOI: 10.3389/fmicb.2022.1071278
Glutamine potentiates gentamicin to kill lab-evolved gentamicin-resistant and clinically isolated multidrug-resistant Escherichia coli
Abstract
Introduction: Gentamicin is a conventional antibiotic in clinic. However, with the wide use of antibiotics, gentamicin-resistant Escherichia coli (E. coli) is an ever-increasing problem that causes infection in both humans and animals. Thus, it is especially important to restore gentamicin-mediated killing efficacy.
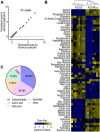
Method: E. coli K12 BW25113 cells were passaged in medium with and without gentamicin and obtain gentamicin-resistant (K12-R GEN ) and control (K12-S) strains, respectively. Then, the metabonomics of the two strains were analyzed by GC-MS approach.
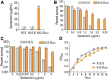
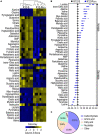
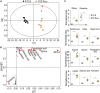
Results: K12-R GEN metabolome was characterized as more decreased metabolites than increased metabolites. Meantime, in the most enriched metabolic pathways, almost all of the metabolites were depressed. Alanine, aspartate and glutamate metabolism and glutamine within the metabolic pathway were identified as the most key metabolic pathways and the most crucial biomarkers, respectively. Exogenous glutamine potentiated gentamicin-mediated killing efficacy in glutamine and gentamicin dose-and time-dependent manners in K12-R GEN . Further experiments showed that glutamine-enabled killing by gentamicin was effective to clinically isolated multidrug-resistant E. coli.
Discussion: These results suggest that glutamine provides an ideal metabolic environment to restore gentamicin-mediated killing, which not only indicates that glutamine is a broad-spectrum antibiotic synergist, but also expands the range of metabolites that contribute to the bactericidal efficiency of aminoglycosides.
Keywords: Escherichia coli; aminoglycoside; antibiotic resistance; glutamine; multidrug resistance; reprogramming metabolomics.
Copyright © 2022 Chen, Ma, Peng and Li.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


References
-
- CLSI (2012). Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria that Grow Aerobically; Approved Standard. CLSI document M07-A9, 9 Edn. Wayne, PA: Clinical and Laboratory Standards Institute.
LinkOut - more resources
Full Text Sources
Miscellaneous

