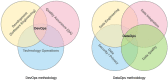
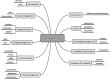
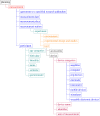
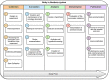
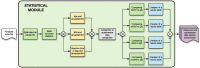
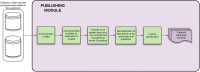
Workflow for health-related and brain data lifecycle
- PMID: 36532611
- PMCID: PMC9748096
- DOI: 10.3389/fdgth.2022.1025086
Workflow for health-related and brain data lifecycle
Abstract
Poor lifestyle leads potentially to chronic diseases and low-grade physical and mental fitness. However, ahead of time, we can measure and analyze multiple aspects of physical and mental health, such as body parameters, health risk factors, degrees of motivation, and the overall willingness to change the current lifestyle. In conjunction with data representing human brain activity, we can obtain and identify human health problems resulting from a long-term lifestyle more precisely and, where appropriate, improve the quality and length of human life. Currently, brain and physical health-related data are not commonly collected and evaluated together. However, doing that is supposed to be an interesting and viable concept, especially when followed by a more detailed definition and description of their whole processing lifecycle. Moreover, when best practices are used to store, annotate, analyze, and evaluate such data collections, the necessary infrastructure development and more intense cooperation among scientific teams and laboratories are facilitated. This approach also improves the reproducibility of experimental work. As a result, large collections of physical and brain health-related data could provide a robust basis for better interpretation of a person's overall health. This work aims to overview and reflect some best practices used within global communities to ensure the reproducibility of experiments, collected datasets and related workflows. These best practices concern, e.g., data lifecycle models, FAIR principles, and definitions and implementations of terminologies and ontologies. Then, an example of how an automated workflow system could be created to support the collection, annotation, storage, analysis, and publication of findings is shown. The Body in Numbers pilot system, also utilizing software engineering best practices, was developed to implement the concept of such an automated workflow system. It is unique just due to the combination of the processing and evaluation of physical and brain (electrophysiological) data. Its implementation is explored in greater detail, and opportunities to use the gained findings and results throughout various application domains are discussed.
Keywords: best practices; brain data; data lifecycle; health information system; health-related data; ontology; physical data; workflow.
© 2022 Brůha, Mouček, Salamon and Vacek.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures







Similar articles
-
The future of Cochrane Neonatal.Early Hum Dev. 2020 Nov;150:105191. doi: 10.1016/j.earlhumdev.2020.105191. Epub 2020 Sep 12. Early Hum Dev. 2020. PMID: 33036834
-
Dietary glycation compounds - implications for human health.Crit Rev Toxicol. 2024 Sep;54(8):485-617. doi: 10.1080/10408444.2024.2362985. Epub 2024 Aug 16. Crit Rev Toxicol. 2024. PMID: 39150724
-
The Effectiveness of Integrated Care Pathways for Adults and Children in Health Care Settings: A Systematic Review.JBI Libr Syst Rev. 2009;7(3):80-129. doi: 10.11124/01938924-200907030-00001. JBI Libr Syst Rev. 2009. PMID: 27820426
-
Critical Care Network in the State of Qatar.Qatar Med J. 2019 Nov 7;2019(2):2. doi: 10.5339/qmj.2019.qccc.2. eCollection 2019. Qatar Med J. 2019. PMID: 31763205 Free PMC article.
-
Impact of summer programmes on the outcomes of disadvantaged or 'at risk' young people: A systematic review.Campbell Syst Rev. 2024 Jun 13;20(2):e1406. doi: 10.1002/cl2.1406. eCollection 2024 Jun. Campbell Syst Rev. 2024. PMID: 38873396 Free PMC article. Review.
References
-
- Salem R, Elsharkawy B, Kader HA. An ontology-based framework for linking heterogeneous medical data. International Conference on Advanced Intelligent Systems, Informatics. Cham (Switzerland): Springer (2016). p. 836–45.
-
- Mavrogiorgou A, Kiourtis A, Kyriazis D. A plug “n” play approach for dynamic data acquisition from heterogeneous IoT medical devices of unknown nature. Evol Syst. (2020) 11:269–89. 10.1007/s12530-019-09286-5 - DOI
-
- Adel E, El-Sappagh S, Barakat S, Elmogy M. Ontology-based electronic health record semantic interoperability: a survey. U-healthcare monitoring systems. Amsterdam: Elsevier (2019). p. 315–52.
Publication types
LinkOut - more resources
Full Text Sources
Research Materials

