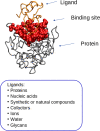
Protein binding sites for drug design
- PMID: 36532870
- PMCID: PMC9734416
- DOI: 10.1007/s12551-022-01028-3
Protein binding sites for drug design
Abstract
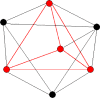
Drug development is a lengthy and challenging process that can be accelerated at early stages by new mathematical approaches and modern computers. To address this important issue, we are developing new mathematical solutions for the detection and characterization of protein binding sites that are important for new drug development. In this review, we present algorithms based on graph theory combined with molecular dynamics simulations that we have developed for studying biological target proteins to provide important data for optimizing the early stages of new drug development. A particular focus is the development of new protein binding site prediction algorithms (ProBiS) and new web tools for modeling pharmaceutically interesting molecules-ProBiS Tools (algorithm, database, web server), which have evolved into a full-fledged graphical tool for studying proteins in the proteome. ProBiS differs from other structural algorithms in that it can align proteins with different folds without prior knowledge of the binding sites. It allows detection of similar binding sites and can predict molecular ligands of various types of pharmaceutical interest that could be advanced to drugs to treat a disease, based on the entire Protein Data Bank (PDB) and AlphaFold database, including proteins not yet in the PDB. All ProBiS Tools are freely available to the academic community at http://insilab.org and https://probis.nih.gov.
Keywords: Prediction; ProBiS; Protein binding sites; Structural proteome.
© International Union for Pure and Applied Biophysics (IUPAB) and Springer-Verlag GmbH Germany, part of Springer Nature 2022, Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
Conflict of interest statement
Conflict of interestThe authors declare no competing interests.
Figures

References
Publication types
LinkOut - more resources
Full Text Sources

