Proteomics analysis of the p.G849D variant in neurexin 2 alpha may reveal insight into Parkinson's disease pathobiology
- PMID: 36533174
- PMCID: PMC9748613
- DOI: 10.3389/fnagi.2022.1002777
Proteomics analysis of the p.G849D variant in neurexin 2 alpha may reveal insight into Parkinson's disease pathobiology
Abstract
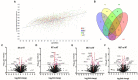
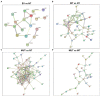
Parkinson's disease (PD), the fastest-growing neurological disorder globally, has a complex etiology. A previous study by our group identified the p.G849D variant in neurexin 2 (NRXN2), encoding the synaptic protein, NRXN2α, as a possible causal variant of PD. Therefore, we aimed to perform functional studies using proteomics in an attempt to understand the biological pathways affected by the variant. We hypothesized that this may reveal insight into the pathobiology of PD. Wild-type and mutant NRXN2α plasmids were transfected into SH-SY5Y cells. Thereafter, total protein was extracted and prepared for mass spectrometry using a Thermo Scientific Fusion mass spectrometer equipped with a Nanospray Flex ionization source. The data were then interrogated against the UniProt H. sapiens database and afterward, pathway and enrichment analyses were performed using in silico tools. Overexpression of the wild-type protein led to the enrichment of proteins involved in neurodegenerative diseases, while overexpression of the mutant protein led to the decline of proteins involved in ribosomal functioning. Thus, we concluded that the wild-type NRXN2α may be involved in pathways related to the development of neurodegenerative disorders, and that biological processes related to the ribosome, transcription, and tRNA, specifically at the synapse, could be an important mechanism in PD. Future studies targeting translation at the synapse in PD could therefore provide further information on the pathobiology of the disease.
Keywords: Parkinson’s disease; mass spectrometry; mitochondrial dysfunction; neurexin 2α (NRXN2); p.G849D; proteomics; ribosomal functioning; synaptic translation.
Copyright © 2022 Cuttler, Fortuin, Müller-Nedebock, Vlok, Cloete and Bardien.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
References
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

