Adaptive exhaustion during prolonged intermittent hypoxia causes dysregulated skeletal muscle protein homeostasis
- PMID: 36533558
- PMCID: PMC10286804
- DOI: 10.1113/JP283700
Adaptive exhaustion during prolonged intermittent hypoxia causes dysregulated skeletal muscle protein homeostasis
Abstract
Nocturnal hypoxaemia, which is common in chronic obstructive pulmonary disease (COPD) patients, is associated with skeletal muscle loss or sarcopenia, which contributes to adverse clinical outcomes. In COPD, we have defined this as prolonged intermittent hypoxia (PIH) because the duration of hypoxia in skeletal muscle occurs through the duration of sleep followed by normoxia during the day, in contrast to recurrent brief hypoxic episodes during obstructive sleep apnoea (OSA). Adaptive cellular responses to PIH are not known. Responses to PIH induced by three cycles of 8 h hypoxia followed by 16 h normoxia were compared to those during chronic hypoxia (CH) or normoxia for 72 h in murine C2C12 and human inducible pluripotent stem cell-derived differentiated myotubes. RNA sequencing followed by downstream analyses were complemented by experimental validation of responses that included both unique and shared perturbations in ribosomal and mitochondrial function during PIH and CH. A sarcopenic phenotype characterized by decreased myotube diameter and protein synthesis, and increased phosphorylation of eIF2α (Ser51) by eIF2α kinase, and of GCN-2 (general controlled non-derepressed-2), occurred during both PIH and CH. Mitochondrial oxidative dysfunction, disrupted supercomplex assembly, lower activity of Complexes I, III, IV and V, and reduced intermediary metabolite concentrations occurred during PIH and CH. Decreased mitochondrial fission occurred during CH. Physiological relevance was established in skeletal muscle of mice with COPD that had increased phosphorylation of eIF2α, lower protein synthesis and mitochondrial oxidative dysfunction. Molecular and metabolic responses with PIH suggest an adaptive exhaustion with failure to restore homeostasis during normoxia. KEY POINTS: Sarcopenia or skeletal muscle loss is one of the most frequent complications that contributes to mortality and morbidity in patients with chronic obstructive pulmonary disease (COPD). Unlike chronic hypoxia, prolonged intermittent hypoxia is a frequent, underappreciated and clinically relevant model of hypoxia in patients with COPD. We developed a novel, in vitro myotube model of prolonged intermittent hypoxia with molecular and metabolic perturbations, mitochondrial oxidative dysfunction, and consequent sarcopenic phenotype. In vivo studies in skeletal muscle from a mouse model of COPD shared responses with our myotube model, establishing the pathophysiological relevance of our studies. These data lay the foundation for translational studies in human COPD to target prolonged, nocturnal hypoxaemia to prevent sarcopenia in these patients.
Keywords: RNA sequencing; intermediary metabolites; mitochondrial oxidation; prolonged intermittent hypoxia; unbiased data.
© 2022 The Authors. The Journal of Physiology © 2022 The Physiological Society.
Conflict of interest statement
Competing interests. All authors disclose that they have no competing interests in accordance with journal policy.
No other conflicts of interest
Figures
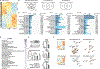

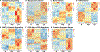
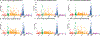

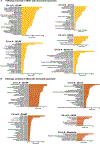


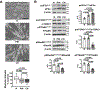




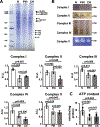


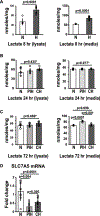

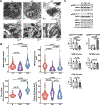

Comment in
-
Muscle cells on a tight budget: cutting expenses during hypoxia.J Physiol. 2023 Feb;601(3):395-396. doi: 10.1113/JP284240. Epub 2023 Jan 19. J Physiol. 2023. PMID: 36627842 No abstract available.
-
Sarcopenia in chronic obstructive pulmonary disease: skeletal muscle gasping for air?J Physiol. 2023 Mar;601(5):883-885. doi: 10.1113/JP284297. Epub 2023 Feb 7. J Physiol. 2023. PMID: 36692139 No abstract available.
References
-
- Barnes PJ. (2016). Sex Differences in Chronic Obstructive Pulmonary Disease Mechanisms. Am J Respir Crit Care Med 193, 813–814. - PubMed
Publication types
MeSH terms
Grants and funding
- RO1 DK113196/NH/NIH HHS/United States
- R56 HL141744/HL/NHLBI NIH HHS/United States
- U01 DK061732/DK/NIDDK NIH HHS/United States
- R01 AA028190/AA/NIAAA NIH HHS/United States
- R21 AA022742/AA/NIAAA NIH HHS/United States
- P50 AA024333/NH/NIH HHS/United States
- R56HL141744;UO1 DK061732/NH/NIH HHS/United States
- U01 AA026976/AA/NIAAA NIH HHS/United States
- 3U01AA026976 - 03S1/NH/NIH HHS/United States
- R21 AR 071046/NH/NIH HHS/United States
- P50 AA024333/AA/NIAAA NIH HHS/United States
- R01 HL155064/HL/NHLBI NIH HHS/United States
- RO1 AA021890/NH/NIH HHS/United States
- U01 DK062470/DK/NIDDK NIH HHS/United States
- UO1 AA 026976/NH/NIH HHS/United States
- R01 GM119174/GM/NIGMS NIH HHS/United States
- R21 AR071046/AR/NIAMS NIH HHS/United States
- K12 HL141952/HL/NHLBI NIH HHS/United States
- 5U01DK062470-17S2/NH/NIH HHS/United States
- R01 DK083414/DK/NIDDK NIH HHS/United States
- U01 AA021890/AA/NIAAA NIH HHS/United States
- R01 CA208516/CA/NCI NIH HHS/United States
- K08 AA028794/AA/NIAAA NIH HHS/United States
- RO1 GM119174/NH/NIH HHS/United States
- R01 DK113196/DK/NIDDK NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical

