Bifunctional Epimerase/Reductase Enzymes Facilitate the Modulation of 6-Deoxy-Heptoses Found in the Capsular Polysaccharides of Campylobacter jejuni
- PMID: 36534477
- PMCID: PMC9838653
- DOI: 10.1021/acs.biochem.2c00633
Bifunctional Epimerase/Reductase Enzymes Facilitate the Modulation of 6-Deoxy-Heptoses Found in the Capsular Polysaccharides of Campylobacter jejuni
Abstract
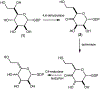
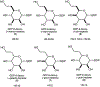
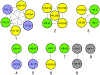
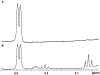
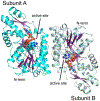
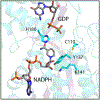
Campylobacter jejuni is a human pathogen and the leading cause of food poisoning in the United States and Europe. Surrounding the exterior surface of this bacterium is a capsular polysaccharide (CPS) that consists of a repeating sequence of common and unusual carbohydrate segments. At least 10 different heptose sugars have thus far been identified in the various strains of C. jejuni. The accepted biosynthetic pathway for the construction of the 6-deoxy-heptoses begins with the 4,6-dehydration of GDP-d-glycero-d-manno-heptose by a dehydratase, followed by an epimerase that racemizes C3 and/or C5 of the product GDP-6-deoxy-4-keto-d-lyxo-heptose. In the final step, a C4-reductase catalyzes the NADPH reduction of the resulting 4-keto product. However, in some strains and serotypes of C. jejuni, there are two separate C4-reductases with different product specificities in the gene cluster for CPS formation. Five pairs of these tandem C4-reductases were isolated, and the catalytic properties were ascertained. In four out of five cases, one of the two C4-reductases is able to catalyze the isomerization of C3 and C5 of GDP-6-deoxy-4-keto-d-lyxo-heptose, in addition to the catalysis of the reduction of C4, thus bypassing the requirement for a separate C3/C5-isomerase. In each case, the 3'-end of the gene for the first C4-reductase contains a poly-G tract of 8-10 guanine residues that may be used to control the expression and/or catalytic activity of either C4-reductase. The three-dimensional structure of the C4-reductase from serotype HS:15, which only does a reduction of C4, was determined to 1.45 Å resolution in the presence of NADPH and GDP.
Conflict of interest statement
The authors declare no competing conflicts of interest.
Figures
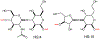

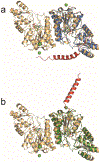
References
-
- Heimesaat MM; Backert S; Alter T; Bereswill S Human Campylobacteriosis-A Serious Infectious Threat in a One Health Perspective. Curr. Top. Microbiol. Immunol 2021, 431, 1–23. - PubMed
-
- Burnham PM; Hendrixson DR Campylobacter jejuni: Collective Components Promoting a Successful Enteric Lifestyle. Nat. Rev. Microbiol 2018, 16, 551–565. - PubMed
-
- Monteiro MA; Noll A; Laird RM; Pequegnat B; Ma ZC; Bertolo L; DePass C; Omari E; Gabryelski P; Redkyna O; Jiao YN; Borrelli S; Poly F; Guerry P Campylobacter jejuni Capsule Polysaccharide Conjugate Vaccine. In Carbohydrate-based Vaccines: from Concept to Clinic; American Chemical Society: Washington, DC, 2018, pp 249–271.
-
- Michael F; Szymanski CM; Li JJ; Chan KH; Khieu NH; Larocque S; Wakarchuk WW; Brisson JR; Monteiro MA The Structures of the Lipooligosaccharide and Capsule Colysaccharide of Campylobacter jejuni Genome Sequenced Strain NCTC 11168. Eur. J. Biochem 2002, 269, 5119–5136. - PubMed
-
- Aspinall GO; Monteiro MA; Pang H Lipo-oligosaccharide of the Campylobacter lari type strain ATCC 35221. Structure of the liberated oligosaccharide and an associated extracellular polysaccharide. Carbohydrate Research 1995, 279, 245–264. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

