Multi-color live-cell STED nanoscopy of mitochondria with a gentle inner membrane stain
- PMID: 36534799
- PMCID: PMC9907107
- DOI: 10.1073/pnas.2215799119
Multi-color live-cell STED nanoscopy of mitochondria with a gentle inner membrane stain
Abstract
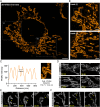
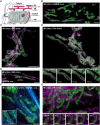
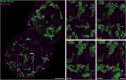
Capturing mitochondria's intricate and dynamic structure poses a daunting challenge for optical nanoscopy. Different labeling strategies have been demonstrated for live-cell stimulated emission depletion (STED) microscopy of mitochondria, but orthogonal strategies are yet to be established, and image acquisition has suffered either from photodamage to the organelles or from rapid photobleaching. Therefore, live-cell nanoscopy of mitochondria has been largely restricted to two-dimensional (2D) single-color recordings of cancer cells. Here, by conjugation of cyclooctatetraene (COT) to a benzo-fused cyanine dye, we report a mitochondrial inner membrane (IM) fluorescent marker, PK Mito Orange (PKMO), featuring efficient STED at 775 nm, strong photostability, and markedly reduced phototoxicity. PKMO enables super-resolution (SR) recordings of IM dynamics for extended periods in immortalized mammalian cell lines, primary cells, and organoids. Photostability and reduced phototoxicity of PKMO open the door to live-cell three-dimensional (3D) STED nanoscopy of mitochondria for 3D analysis of the convoluted IM. PKMO is optically orthogonal with green and far-red markers, allowing multiplexed recordings of mitochondria using commercial STED microscopes. Using multi-color STED microscopy, we demonstrate that imaging with PKMO can capture interactions of mitochondria with different cellular components such as the endoplasmic reticulum (ER) or the cytoskeleton, Bcl-2-associated X protein (BAX)-induced apoptotic process, or crista phenotypes in genetically modified cells, all at sub-100 nm resolution. Thereby, this work offers a versatile tool for studying mitochondrial IM architecture and dynamics in a multiplexed manner.
Keywords: STED nanoscopy; cristae; mitochondria.
Conflict of interest statement
The authors declare competing interest. The authors have organizational affiliations to disclose, Z.C. is an inventor of the patent on the mitochondria dye described in this work (CN 202010492298.8). The patent was applied through Peking University and is currently transferred to Genvivo Tech (in which Z.C. is a founder and shareholder) for commercialization.
Figures




References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

