Endothelial PTP4A1 mitigates vascular inflammation via USF1/A20 axis-mediated NF-κB inactivation
- PMID: 36534975
- PMCID: PMC10411943
- DOI: 10.1093/cvr/cvac193
Endothelial PTP4A1 mitigates vascular inflammation via USF1/A20 axis-mediated NF-κB inactivation
Abstract
Aims: The nuclear factor-κB (NF-κB) signalling pathway plays a critical role in the pathogenesis of multiple vascular diseases. However, in endothelial cells (ECs), the molecular mechanisms responsible for the negative regulation of the NF-κB pathway are poorly understood. In this study, we investigated a novel role for protein tyrosine phosphatase type IVA1 (PTP4A1) in NF-κB signalling in ECs.
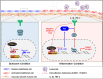
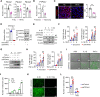
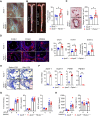
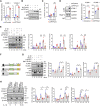
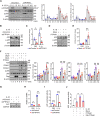
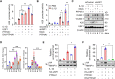
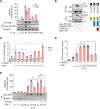
Methods and results: In human tissues, human umbilical artery ECs, and mouse models for loss of function and gain of function of PTP4A1, we conducted histological analysis, immunostaining, laser-captured microdissection assay, lentiviral infection, small interfering RNA transfection, quantitative real-time PCR and reverse transcription-PCR, as well as luciferase reporter gene and chromatin immunoprecipitation assays. Short hairpin RNA-mediated knockdown of PTP4A1 and overexpression of PTP4A1 in ECs indicated that PTP4A1 is critical for inhibiting the expression of cell adhesion molecules (CAMs). PTP4A1 increased the transcriptional activity of upstream stimulatory factor 1 (USF1) by dephosphorylating its S309 residue and subsequently inducing the transcription of tumour necrosis factor-alpha-induced protein 3 (TNFAIP3/A20) and the inhibition of NF-κB activity. Studies on Ptp4a1 knockout or transgenic mice demonstrated that PTP4A1 potently regulates the interleukin 1β-induced expression of CAMs in vivo. In addition, we verified that PTP4A1 deficiency in apolipoprotein E knockout mice exacerbated high-fat high-cholesterol diet-induced atherogenesis with upregulated expression of CAMs.
Conclusion: Our data indicate that PTP4A1 is a novel negative regulator of vascular inflammation by inducing USF1/A20 axis-mediated NF-κB inactivation. Therefore, the expression and/or activation of PTP4A1 in ECs might be useful for the treatment of vascular inflammatory diseases.
Keywords: Atherosclerosis; Cell adhesion molecules; Endothelial cells; Protein phosphatase; Vascular inflammation.
© The Author(s) 2022. Published by Oxford University Press on behalf of the European Society of Cardiology. All rights reserved. For permissions, please e-mail: journals.permissions@oup.com.
Conflict of interest statement
Conflict of interest: None declared.
Figures

References
-
- Ley K, Laudanna C, Cybulsky MI, Nourshargh S. Getting to the site of inflammation: the leukocyte adhesion cascade updated. Nat Rev Immunol 2007;7:678–689. - PubMed
-
- Cotran RS, Mayadas-Norton T. Endothelial adhesion molecules in health and disease. Pathol Biol 1998;46:164–170. - PubMed
-
- Park JG, Ryu SY, Jung IH, Lee YH, Kang KJ, Lee MR, Lee MN, Sonn SK, Lee JH, Lee H, Oh GT, Moon K, Shim H. Evaluation of VCAM-1 antibodies as therapeutic agent for atherosclerosis in apolipoprotein E-deficient mice. Atherosclerosis 2013;226:356–363. - PubMed
-
- Kitagawa K, Matsumoto M, Sasaki T, Hashimoto H, Kuwabara K, Ohtsuki T, Hori M. Involvement of ICAM-1 in the progression of atherosclerosis in APOE-knockout mice. Atherosclerosis 2002;160:305–310. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

