The tongue biofilm metatranscriptome identifies metabolic pathways associated with the presence or absence of halitosis
- PMID: 36535943
- PMCID: PMC9763428
- DOI: 10.1038/s41522-022-00364-2
The tongue biofilm metatranscriptome identifies metabolic pathways associated with the presence or absence of halitosis
Abstract
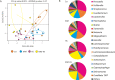
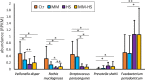
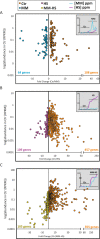
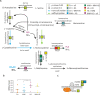
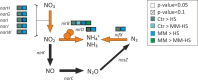
Intra-oral halitosis usually results from the production of volatile sulfur compounds, such as methyl mercaptan and hydrogen sulfide, by the tongue microbiota. There are currently no reports on the microbial gene-expression profiles of the tongue microbiota in halitosis. In this study, we performed RNAseq of tongue coating samples from individuals with and without halitosis. The activity of Streptococcus (including S. parasanguinis), Veillonella (including V. dispar) and Rothia (including R. mucilaginosa) was associated with halitosis-free individuals while Prevotella (including P. shahi), Fusobacterium (including F. nucleatum) and Leptotrichia were associated with halitosis. Interestingly, the metatranscriptome of patients that only had halitosis levels of methyl mercaptan was similar to that of halitosis-free individuals. Finally, gene expression profiles showed a significant over-expression of genes involved in L-cysteine and L-homocysteine synthesis, as well as nitrate reduction genes, in halitosis-free individuals and an over-expression of genes responsible for cysteine degradation into hydrogen sulfide in halitosis patients.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

