Adaptive immune responses to SARS-CoV-2 persist in the pharyngeal lymphoid tissue of children
- PMID: 36536106
- PMCID: PMC10777159
- DOI: 10.1038/s41590-022-01367-z
Adaptive immune responses to SARS-CoV-2 persist in the pharyngeal lymphoid tissue of children
Abstract
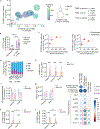
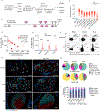
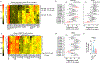
Most studies of adaptive immunity to SARS-CoV-2 infection focus on peripheral blood, which may not fully reflect immune responses at the site of infection. Using samples from 110 children undergoing tonsillectomy and adenoidectomy during the COVID-19 pandemic, we identified 24 samples with evidence of previous SARS-CoV-2 infection, including neutralizing antibodies in serum and SARS-CoV-2-specific germinal center and memory B cells in the tonsils and adenoids. Single-cell B cell receptor (BCR) sequencing indicated virus-specific BCRs were class-switched and somatically hypermutated, with overlapping clones in the two tissues. Expanded T cell clonotypes were found in tonsils, adenoids and blood post-COVID-19, some with CDR3 sequences identical to previously reported SARS-CoV-2-reactive T cell receptors (TCRs). Pharyngeal tissues from COVID-19-convalescent children showed persistent expansion of germinal center and antiviral lymphocyte populations associated with interferon (IFN)-γ-type responses, particularly in the adenoids, and viral RNA in both tissues. Our results provide evidence for persistent tissue-specific immunity to SARS-CoV-2 in the upper respiratory tract of children after infection.
© 2022. This is a U.S. Government work and not under copyright protection in the US; foreign copyright protection may apply.
Conflict of interest statement
Figures















Update of
-
Robust, persistent adaptive immune responses to SARS-CoV-2 in the oropharyngeal lymphoid tissue of children.Res Sq [Preprint]. 2022 Mar 23:rs.3.rs-1276578. doi: 10.21203/rs.3.rs-1276578/v1. Res Sq. 2022. Update in: Nat Immunol. 2023 Jan;24(1):186-199. doi: 10.1038/s41590-022-01367-z. PMID: 35350206 Free PMC article. Updated. Preprint.
References
Methods References:
-
- Zahra FT, Bellusci L, Grubbs G, Golding H & Khurana S Neutralisation of circulating SARS-CoV-2 delta and omicron variants by convalescent plasma and SARS-CoV-2 hyperimmune intravenous human immunoglobulins for treatment of COVID-19. Annals of the Rheumatic Diseases, annrheumdis-2022–222115, doi:10.1136/annrheumdis-2022-222115 (2022). - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

