Liver RBFOX2 regulates cholesterol homeostasis via Scarb1 alternative splicing in mice
- PMID: 36536133
- PMCID: PMC9771820
- DOI: 10.1038/s42255-022-00681-y
Liver RBFOX2 regulates cholesterol homeostasis via Scarb1 alternative splicing in mice
Abstract
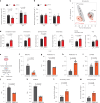
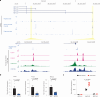
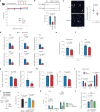
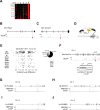
RNA alternative splicing (AS) expands the regulatory potential of eukaryotic genomes. The mechanisms regulating liver-specific AS profiles and their contribution to liver function are poorly understood. Here, we identify a key role for the splicing factor RNA-binding Fox protein 2 (RBFOX2) in maintaining cholesterol homeostasis in a lipogenic environment in the liver. Using enhanced individual-nucleotide-resolution ultra-violet cross-linking and immunoprecipitation, we identify physiologically relevant targets of RBFOX2 in mouse liver, including the scavenger receptor class B type I (Scarb1). RBFOX2 function is decreased in the liver in diet-induced obesity, causing a Scarb1 isoform switch and alteration of hepatocyte lipid homeostasis. Our findings demonstrate that specific AS programmes actively maintain liver physiology, and underlie the lipotoxic effects of obesogenic diets when dysregulated. Splice-switching oligonucleotides targeting this network alleviate obesity-induced inflammation in the liver and promote an anti-atherogenic lipoprotein profile in the blood, underscoring the potential of isoform-specific RNA therapeutics for treating metabolism-associated diseases.
© 2022. The Author(s).
Conflict of interest statement
C.R.S. is inventor on a patent covering the eiCLIP method that has been filed in the UK (2006803.7) and internationally (PCT/GB2021/051109). The other authors declare no competing interests.
Figures






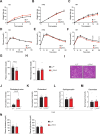
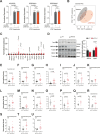
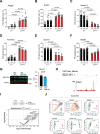

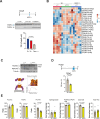

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

