mTORC1 regulates a lysosome-dependent adaptive shift in intracellular lipid species
- PMID: 36536136
- PMCID: PMC9799240
- DOI: 10.1038/s42255-022-00706-6
mTORC1 regulates a lysosome-dependent adaptive shift in intracellular lipid species
Abstract
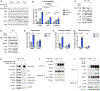
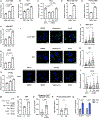
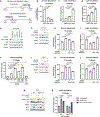
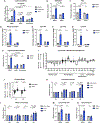
The mechanistic target of rapamycin complex 1 (mTORC1) senses and relays environmental signals from growth factors and nutrients to metabolic networks and adaptive cellular systems to control the synthesis and breakdown of macromolecules; however, beyond inducing de novo lipid synthesis, the role of mTORC1 in controlling cellular lipid content remains poorly understood. Here we show that inhibition of mTORC1 via small molecule inhibitors or nutrient deprivation leads to the accumulation of intracellular triglycerides in both cultured cells and a mouse tumor model. The elevated triglyceride pool following mTORC1 inhibition stems from the lysosome-dependent, but autophagy-independent, hydrolysis of phospholipid fatty acids. The liberated fatty acids are available for either triglyceride synthesis or β-oxidation. Distinct from the established role of mTORC1 activation in promoting de novo lipid synthesis, our data indicate that mTORC1 inhibition triggers membrane phospholipid trafficking to the lysosome for catabolism and an adaptive shift in the use of constituent fatty acids for storage or energy production.
© 2022. The Author(s), under exclusive licence to Springer Nature Limited.
Conflict of interest statement
Declaration of interests
B.D.M. is a member of the scientific advisory board and a shareholder of Navitor Pharmaceuticals. All other authors declare no competing financial interests.
Figures






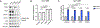






Comment in
-
mTORC1 takes control of lysosomal lipid breakdown.Nat Metab. 2022 Dec;4(12):1620-1622. doi: 10.1038/s42255-022-00702-w. Nat Metab. 2022. PMID: 36536135 No abstract available.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

