Multi-omics integration reveals a core network involved in host defence and hyperkeratinization in psoriasis
- PMID: 36536476
- PMCID: PMC9763538
- DOI: 10.1002/ctm2.976
Multi-omics integration reveals a core network involved in host defence and hyperkeratinization in psoriasis
Abstract
Objectives: The precise pathogenesis of psoriasis remains incompletely explored. We aimed to better understand the underlying mechanisms of psoriasis, using a systems biology approach based on transcriptomics and microbiome profiling.
Methods: We collected the skin tissue biopsies and swabs in both lesional and non-lesional skin of 13 patients with psoriasis, 15 patients with psoriatic arthritis and healthy skin from 12 patients with ankylosing spondylitis. To study the similarities and differences in the molecular profiles between these three conditions, and the associations between the host defence and microbiota composition, we performed high-throughput RNA-sequencing to quantify the gene expression profile in tissues. The metagenomic composition of 16S on local skin sites was quantified by clustering amplicon sequences and counted into operational taxonomic units. We further analysed associations between the transcriptome and microbiome profiling.
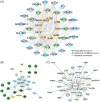
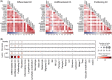
Results: We found that lesional and non-lesional samples were remarkably different in terms of their transcriptome profiles. The functional annotation of differentially expressed genes showed a major enrichment in neutrophil activation. By using co-expression gene networks, we identified a gene module that was associated with local psoriasis severity at the site of biopsy. From this module, we found a 'core' set of genes that was functionally involved in neutrophil activation, epidermal cell differentiation and response to bacteria. Skin microbiome analysis revealed that the abundances of Enhydrobacter, Micrococcus and Leptotrichia were significantly correlated with the genes in core network.
Conclusions: We identified a core gene network that associated with local disease severity and microbiome composition, involved in the inflammation and hyperkeratinization in psoriatic skin.
Keywords: gene network; hyperkeratinization; multi-omics; psoriasis.
© 2022 The Authors. Clinical and Translational Medicine published by John Wiley & Sons Australia, Ltd on behalf of Shanghai Institute of Clinical Bioinformatics.
Conflict of interest statement
T.R. received consultancy fees from Jansen in 2016 and 2017 on topics that were unrelated to the content of this manuscript. T.R., A.P., and W.T. are currently an employee of AbbVie, with no conflicts of interest regarding the work of this manuscript. D.B. received consultancy fees from Janssen in 2018 and 2019 on topics that were unrelated to the content of this manuscript. The other authors have declared no conflicts of interest.
Figures




Similar articles
-
Exploratory multi-omics analysis reveals host-microbe interactions associated with disease severity in psoriatic skin.EBioMedicine. 2024 Jul;105:105222. doi: 10.1016/j.ebiom.2024.105222. Epub 2024 Jun 25. EBioMedicine. 2024. PMID: 38924840 Free PMC article.
-
Comparisons of gene expression in normal, lesional, and non-lesional psoriatic skin using DNA microarray techniques.Int J Dermatol. 2014 Oct;53(10):1213-20. doi: 10.1111/ijd.12476. Epub 2014 Jul 11. Int J Dermatol. 2014. PMID: 25041445
-
Transcriptional landscape of psoriasis identifies the involvement of IL36 and IL36RN.BMC Genomics. 2015 Apr 19;16(1):322. doi: 10.1186/s12864-015-1508-2. BMC Genomics. 2015. PMID: 25897967 Free PMC article.
-
Transcriptional Basis of Psoriasis from Large Scale Gene Expression Studies: The Importance of Moving towards a Precision Medicine Approach.Int J Mol Sci. 2022 May 30;23(11):6130. doi: 10.3390/ijms23116130. Int J Mol Sci. 2022. PMID: 35682804 Free PMC article. Review.
-
MicroRNA profiling of psoriatic skin identifies 11 miRNAs associated with disease severity.Exp Dermatol. 2022 Apr;31(4):535-547. doi: 10.1111/exd.14497. Epub 2021 Nov 17. Exp Dermatol. 2022. PMID: 34748247 Review.
Cited by
-
Complement C3 deficient mice show more severe imiquimod-induced psoriasiform dermatitis than wild-type mice regardless of the commensal microbiota.Exp Anim. 2024 Oct 23;73(4):458-467. doi: 10.1538/expanim.24-0043. Epub 2024 Jun 29. Exp Anim. 2024. PMID: 38945882 Free PMC article.
-
CASZ1 Is Essential for Skin Epidermal Terminal Differentiation.J Invest Dermatol. 2024 Sep;144(9):2029-2038. doi: 10.1016/j.jid.2024.02.014. Epub 2024 Mar 6. J Invest Dermatol. 2024. PMID: 38458428 Free PMC article.
-
Injectable smart-blended hydrogel cross-linked with Vanillin to accelerate differentiation of intervertebral disc-derived stem cells (IVDSCs) for promoting degenerative nucleolus pulposus in a rat model.Inflammopharmacology. 2024 Oct;32(5):3443-3459. doi: 10.1007/s10787-024-01554-4. Epub 2024 Aug 29. Inflammopharmacology. 2024. PMID: 39207637
-
Multi-Target Mechanism of Compound Qingdai Capsule for Treatment of Psoriasis: Multi-Omics Analysis and Experimental Verification.Drug Des Devel Ther. 2025 Jun 18;19:5209-5230. doi: 10.2147/DDDT.S523836. eCollection 2025. Drug Des Devel Ther. 2025. PMID: 40546661 Free PMC article.
-
Impact of silencing eEF2K expression on the malignant properties of chordoma.Mol Biol Rep. 2023 Apr;50(4):3011-3022. doi: 10.1007/s11033-023-08257-z. Epub 2023 Jan 18. Mol Biol Rep. 2023. PMID: 36652154
References
-
- Sultana A, Bhuiyan SI, Mahmud MM, Siddique RU, Shawkat SM, Nandi AK. Comorbidities in patients with psoriasis. Mymensingh Med J. 2019;28(4):894‐899. - PubMed
-
- World Health Organization . Global Report on Psoriasis: World Health Organization . Published online 2016. https://apps.who.int/iris/handle/10665/204417
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
