Raman spectroscopic cellomics for the detection of SARS-CoV-2-associated neutrophil activation after TNF-α stimulation
- PMID: 36536489
- PMCID: PMC9763540
- DOI: 10.1002/ctm2.1139
Raman spectroscopic cellomics for the detection of SARS-CoV-2-associated neutrophil activation after TNF-α stimulation
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures

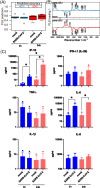
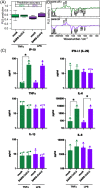
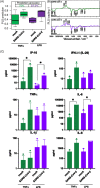
References
-
- Schie IW, Ruger J, Mondol AS, et al. High‐throughput screening Raman spectroscopy platform for label‐free cellomics. Anal Chem. 2018;90:2023‐2030. - PubMed
-
- Arend N, Pittner A, Ramoji A, et al. Detection and differentiation of bacterial and fungal infection of neutrophils from peripheral blood using Raman spectroscopy. Anal Chem. 2020;92:10560‐10568. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous
