Lab-on-a-chip for the easy and visual detection of SARS-CoV-2 in saliva based on sensory polymers
- PMID: 36536612
- PMCID: PMC9751010
- DOI: 10.1016/j.snb.2022.133165
Lab-on-a-chip for the easy and visual detection of SARS-CoV-2 in saliva based on sensory polymers
Abstract
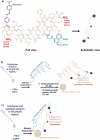
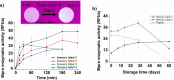
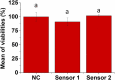
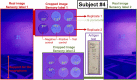
The initial stages of the pandemic caused by SARS-CoV-2 showed that early detection of the virus in a simple way is the best tool until the development of vaccines. Many different tests are invasive or need the patient to cough up or even drag a sample of mucus from the throat area. Besides, the manufacturing time has proven insufficient in pandemic conditions since they were out of stock in many countries. Here we show a new method of manufacturing virus sensors and a proof of concept with SARS-CoV-2. We found that a fluorogenic peptide substrate of the main protease of the virus (Mpro) can be covalently immobilized in a polymer, with which a cellulose-based material can be coated. These sensory labels fluoresce with a single saliva sample of a positive COVID-19 patient. The results matched with that of the antigen tests in 22 of 26 studied cases (85% success rate).
Keywords: COVID-19; Coating; Fluorimetry; Immobilization; Pandemics; Paper-supported; Peptide; Substrate.
© 2022 The Authors.
Conflict of interest statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures

References
-
- University of Oxford, COVID-19 Data Explorer, (2022). 〈https://ourworldindata.org/explorers/coronavirus-data-explorer〉 (Accessed March 7, 2022).
-
- Hale T., Angrist N., Goldszmidt R., Kira B., Petherick A., Phillips T., Webster S., Cameron-Blake E., Hallas L., Majumdar S., Tatlow H. A global panel database of pandemic policies (Oxford COVID-19 Government Response Tracker) Nat. Hum. Behav. 2021;5:529–538. doi: 10.1038/S41562-021-01079-8. - DOI - PubMed
-
- Artik Y., Kurtulmus M.S., Cesur N.P., Mart Komurcu S.Z., Kazezoglu C., Kocatas A. Clinic evaluation of the destrovir spray effectiveness in SARS-CoV-2 disease. Electron. J. Gen. Med. 2022;19:em357. doi: 10.29333/EJGM/11578. - DOI
-
- Fernandes Q., Inchakalody V.P., Merhi M., Mestiri S., Taib N., Moustafa Abo El-Ella D., Bedhiafi T., Raza A., Al-Zaidan L., Mohsen M.O., Yousuf Al-Nesf M.A., Hssain A.A., Yassine H.M., Bachmann M.F., Uddin S., Dermime S. Emerging COVID-19 variants and their impact on SARS-CoV-2 diagnosis, therapeutics and vaccines. Ann. Med. 2022;54:524–540. doi: 10.1080/07853890.2022.2031274. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous
