SARS-CoV-2 multi-variant rapid detector based on graphene transistor functionalized with an engineered dimeric ACE2 receptor
- PMID: 36536857
- PMCID: PMC9750890
- DOI: 10.1016/j.nantod.2022.101729
SARS-CoV-2 multi-variant rapid detector based on graphene transistor functionalized with an engineered dimeric ACE2 receptor
Abstract
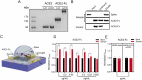
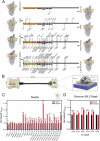
Reliable point-of-care (POC) rapid tests are crucial to detect infection and contain the spread of Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2). The emergence of several variants of concern (VOC) can reduce binding affinity to diagnostic antibodies, limiting the efficacy of the currently adopted tests, while showing unaltered or increased affinity for the host receptor, angiotensin converting enzyme 2 (ACE2). We present a graphene field-effect transistor (gFET) biosensor design, which exploits the Spike-ACE2 interaction, the crucial step for SARS-CoV-2 infection. Extensive computational analyses show that a chimeric ACE2-Fragment crystallizable (ACE2-Fc) construct mimics the native receptor dimeric conformation. ACE2-Fc functionalized gFET allows in vitro detection of the trimeric Spike protein, outperforming functionalization with a diagnostic antibody or with the soluble ACE2 portion, resulting in a sensitivity of 20 pg/mL. Our miniaturized POC biosensor successfully detects B.1.610 (pre-VOC), Alpha, Beta, Gamma, Delta, Omicron (i.e., BA.1, BA.2, BA.4, BA.5, BA.2.75 and BQ.1) variants in isolated viruses and patient's clinical nasopharyngeal swabs. The biosensor reached a Limit Of Detection (LOD) of 65 cps/mL in swab specimens of Omicron BA.5. Our approach paves the way for a new and reusable class of highly sensitive, rapid and variant-robust SARS-CoV-2 detection systems.
Keywords: Biosensor; Centaurus; Cerberus; Molecular dynamics; Omicron; Point-of-care; SARS-CoV-2 variants; gFET.
© 2022 The Authors. Published by Elsevier Ltd.
Conflict of interest statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper. The ACE2-Fc functionalization of gFET and the design of the POC device are under patent pending, applied by Polytechnic University of Marche. D.DM., M.DA., C.A., A.R., I.DA., D.M., E.P., P.C., G.B., L.P., M.F. are the inventors of the patent application N. 102021000000533 filed in 01/13/2021. All other authors declare they have no competing interests.
Figures




References
-
-
(2021). https://outbreak.info/ (accessed September 17, 2021).
-
LinkOut - more resources
Full Text Sources
Miscellaneous
