Molecular Characterization of Aspergillus flavus Strains Isolated from Animal Feeds
- PMID: 36537059
- PMCID: PMC9944975
- DOI: 10.33073/pjm-2022-048
Molecular Characterization of Aspergillus flavus Strains Isolated from Animal Feeds
Abstract
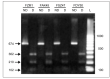
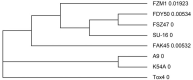
Aflatoxin (AF)-producing fungi such as Aspergillus flavus commonly contaminate animal feeds, causing high economic losses. A. flavus is the most prevalent and produces AFB1, a potent mutagen, and carcinogen threatening human and animal health. Aspergillaceae is a large group of closely related fungi sharing number of morphological and genetic similarities that complicate the diagnosis of highly pathogenic strains. We used here morphological and molecular assays to characterize fungal isolates from animal feeds in Southwestern Algeria. These tools helped to identify 20 out of 30 Aspergillus strains, and 15 of them belonged to the Aspergillus section Flavi. Further analyses detected four out of 15 as belonging to Aspergillus flavus-parasiticus group. PCR targeting the AF genes' aflR-aflS(J) intergenic region amplified a single 674 bp amplicon in all four isolates. The amplicons were digested with a BglII endonuclease, and three specific fragments were observed for A. flavus but A. parasitucus lacked two typical fragments. Sequencing data of four amplicons confirmed the presence of the two BglII restriction sites yielding the three fragments, confirming that all four strains were A. flavus. In addition, this analysis illustrated the genetic variability within the A. flavus strains.
Keywords: Aspergillus flavus; IGS; PCR-RFLP; aflR-aflS(J); diagnosis tools.
© 2022 Hadjer Saber et al., published by Sciendo.
Conflict of interest statement
The authors do not report any financial or personal connections with other persons or organizations, which might negatively affect the contents of this publication and/or claim authorship rights to this publication.
Figures



Similar articles
-
Differentiation between Aspergillus flavus and Aspergillus parasiticus from pure culture and aflatoxin-contaminated grapes using PCR-RFLP analysis of aflR-aflJ intergenic spacer.J Food Sci. 2011 May;76(4):M247-53. doi: 10.1111/j.1750-3841.2011.02153.x. Epub 2011 Apr 14. J Food Sci. 2011. PMID: 22417364
-
[Discrimination of Aspergillus flavus group fungi using phylogenetic tree analysis and multiplex PCR].Shokuhin Eiseigaku Zasshi. 2014;55(3):135-41. doi: 10.3358/shokueishi.55.135. Shokuhin Eiseigaku Zasshi. 2014. PMID: 24990760 Japanese.
-
Isolation and characterization of Aspergillus flavus strains in China.J Microbiol. 2018 Feb;56(2):119-127. doi: 10.1007/s12275-018-7144-1. Epub 2018 Feb 2. J Microbiol. 2018. PMID: 29392555
-
PCR detection of aflatoxin producing fungi and its limitations.Int J Food Microbiol. 2012 May 1;156(1):1-6. doi: 10.1016/j.ijfoodmicro.2012.03.001. Epub 2012 Mar 7. Int J Food Microbiol. 2012. PMID: 22445201 Review.
-
What does genetic diversity of Aspergillus flavus tell us about Aspergillus oryzae?Int J Food Microbiol. 2010 Apr 15;138(3):189-99. doi: 10.1016/j.ijfoodmicro.2010.01.033. Epub 2010 Feb 1. Int J Food Microbiol. 2010. PMID: 20163884 Review.
Cited by
-
Characterization of Aspergillus section Flavi associated with stored grains.Mycotoxin Res. 2024 Feb;40(1):187-202. doi: 10.1007/s12550-023-00514-1. Epub 2024 Jan 17. Mycotoxin Res. 2024. PMID: 38231446 Free PMC article.
-
Aspergillus Mycotoxins: The Major Food Contaminants.Adv Sci (Weinh). 2025 Mar;12(9):e2412757. doi: 10.1002/advs.202412757. Epub 2025 Feb 7. Adv Sci (Weinh). 2025. PMID: 39921319 Free PMC article. Review.
References
-
- Abriba C, Lennox JA, Asikong BE, Asitok A, Ikpoh IS, Henshaw EE, Eja ME. Isolation of aflatoxin producing species of Aspergillus from foodstuffs sold in Calabar markets, Cross River state, Nigeria. J Microbiol Biotechnol Res. 2013;3:8–13.
-
- Ahmad MM, Ahmad M, Ali A, Hamid R, Javed S, Abdin MZ. Gupta VK, Tuohy M, Gaur RK. Fungal biochemistry and biotechnology. Saarbrücken (Germany): Lambert; 2010. Molecular methods for detecting pathogenic fungi; pp. 154–169. p.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
