Heterologous chimpanzee adenovirus vector immunizations for SARS-CoV-2 spike and nucleocapsid protect hamsters against COVID-19
- PMID: 36539010
- PMCID: PMC9758783
- DOI: 10.1016/j.micinf.2022.105082
Heterologous chimpanzee adenovirus vector immunizations for SARS-CoV-2 spike and nucleocapsid protect hamsters against COVID-19
Abstract
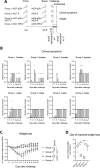
Available COVID-19 vaccine only provide protection for a limited time due in part to the rapid emergence of viral variants with spike protein mutations, necessitating the generation of new vaccines to combat SARS-CoV-2. Two serologically distinct replication-defective chimpanzee-origin adenovirus (Ad) vectors (AdC) called AdC6 and AdC7 expressing early SARS-CoV-2 isolate spike (S) or nucleocapsid (N) proteins, the latter expressed as a fusion protein within herpes simplex virus glycoprotein D (gD), were tested individually or as a mixture in a hamster COVID-19 SARS-CoV-2 challenge model. The S protein expressing AdC (AdC-S) vectors induced antibodies including those with neutralizing activity that in part cross-reacted with viral variants. Hamsters vaccinated with the AdC-S vectors were protected against serious disease and showed accelerated recovery upon SARS-CoV-2 challenge. Protection was enhanced if AdC-S vectors were given together with the AdC vaccines that expressed the gD N fusion protein (AdC-gDN). In contrast hamsters that just received the AdC-gDN vaccines showed only marginal lessening of symptoms compared to control animals. These results indicate that immune response to the N protein that is less variable than the S protein may potentiate and prolong protection achieved by the currently used S protein based genetic COVID-19 vaccines.
Keywords: Adenovirus vector vaccines; COVID-19 vaccine; Hamster challenge model; Nucleocapsid protein; Spike protein.
Copyright © 2022 Institut Pasteur. Published by Elsevier Masson SAS. All rights reserved.
Conflict of interest statement
Declaration of competing interest HCJE holds equity in Virion Therapeutics. She serves as a Consultant to several Gene Therapy companies.
Figures



References
-
- Emary K.R.W., Golubchik T., Aley P.K., Ariani C.V., Angus B., Bibi S., et al. Efficacy of ChAdOx1 nCoV-19 (AZD1222) vaccine against SARS-CoV-2 variant of concern 202012/01 (B.1.1.7): an exploratory analysis of a randomised controlled trial. Lancet. 2021;397:1351–1362. doi: 10.1016/S0140-6736(21)00628-0. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

