Tracking the clonal dynamics of SARS-CoV-2-specific T cells in children and adults with mild/asymptomatic COVID-19
- PMID: 36539107
- PMCID: PMC9758763
- DOI: 10.1016/j.clim.2022.109209
Tracking the clonal dynamics of SARS-CoV-2-specific T cells in children and adults with mild/asymptomatic COVID-19
Abstract
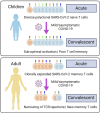
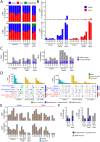
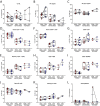
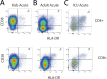
Children infected with severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) develop less severe coronavirus disease 2019 (COVID-19) than adults. The mechanisms for the age-specific differences and the implications for infection-induced immunity are beginning to be uncovered. We show by longitudinal multimodal analysis that SARS-CoV-2 leaves a small footprint in the circulating T cell compartment in children with mild/asymptomatic COVID-19 compared to adult household contacts with the same disease severity who had more evidence of systemic T cell interferon activation, cytotoxicity and exhaustion. Children harbored diverse polyclonal SARS-CoV-2-specific naïve T cells whereas adults harbored clonally expanded SARS-CoV-2-specific memory T cells. A novel population of naïve interferon-activated T cells is expanded in acute COVID-19 and is recruited into the memory compartment during convalescence in adults but not children. This was associated with the development of robust CD4+ memory T cell responses in adults but not children. These data suggest that rapid clearance of SARS-CoV-2 in children may compromise their cellular immunity and ability to resist reinfection.
Keywords: COVID-19; Clonal dynamics; Interferon-activated naive T cells; Memory T cells; SARS-CoV-2; Single cell transcriptomics; T cell receptor repertoire.
Copyright © 2022 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of Competing Interest The authors declare no competing financial interests.
Figures









References
-
- Zhou P., Yang X.L., Wang X.G., Hu B., Zhang L., Zhang W., Si H.R., Zhu Y., Li B., Huang C.L., Chen H.D., Chen J., Luo Y., Guo H., Jiang R.D., Liu M.Q., Chen Y., Shen X.R., Wang X., Zheng X.S., Zhao K., Chen Q.J., Deng F., Liu L.L., Yan B., Zhan F.X., Wang Y.Y., Xiao G.F., Shi Z.L. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. - PMC - PubMed
-
- Guan W.J., Ni Z.Y., Hu Y., Liang W.H., Ou C.Q., He J.X., Liu L., Shan H., Lei C.L., Hui D.S.C., Du B., Li L.J., Zeng G., Yuen K.Y., Chen R.C., Tang C.L., Wang T., Chen P.Y., Xiang J., Li S.Y., Wang J.L., Liang Z.J., Peng Y.X., Wei L., Liu Y., Hu Y.H., Peng P., Wang J.M., Liu J.Y., Chen Z., Li G., Zheng Z.J., Qiu S.Q., Luo J., Ye C.J., Zhu S.Y., Zhong N.S., China Medical Treatment Expert Group for, C Clinical Characteristics of Coronavirus Disease 2019 in China. N. Engl. J. Med. 2020;382:1708–1720. - PMC - PubMed
-
- Chen T., Wu D., Chen H., Yan W., Yang D., Chen G., Ma K., Xu D., Yu H., Wang H., Wang T., Guo W., Chen J., Ding C., Zhang X., Huang J., Han M., Li S., Luo X., Zhao J., Ning Q. Clinical characteristics of 113 deceased patients with coronavirus disease 2019: retrospective study. BMJ. 2020;368 - PMC - PubMed
-
- Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., Zhang L., Fan G., Xu J., Gu X., Cheng Z., Yu T., Xia J., Wei Y., Wu W., Xie X., Yin W., Li H., Liu M., Xiao Y., Gao H., Guo L., Xie J., Wang G., Jiang R., Gao Z., Jin Q., Wang J., Cao B. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

