DPAM: A domain parser for AlphaFold models
- PMID: 36539305
- PMCID: PMC9850437
- DOI: 10.1002/pro.4548
DPAM: A domain parser for AlphaFold models
Abstract
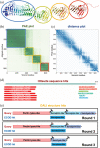
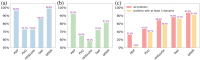
The recent breakthroughs in structure prediction, where methods such as AlphaFold demonstrated near-atomic accuracy, herald a paradigm shift in structural biology. The 200 million high-accuracy models released in the AlphaFold Database are expected to guide protein science in the coming decades. Partitioning these AlphaFold models into domains and assigning them to an evolutionary hierarchy provide an efficient way to gain functional insights into proteins. However, classifying such a large number of predicted structures challenges the infrastructure of current structure classifications, including our Evolutionary Classification of protein Domains (ECOD). Better computational tools are urgently needed to parse and classify domains from AlphaFold models automatically. Here we present a Domain Parser for AlphaFold Models (DPAM) that can automatically recognize globular domains from these models based on inter-residue distances in 3D structures, predicted aligned errors, and ECOD domains found by sequence (HHsuite) and structural (Dali) similarity searches. Based on a benchmark of 18,759 AlphaFold models, we demonstrate that DPAM can recognize 98.8% of domains and assign correct boundaries for 87.5%, significantly outperforming structure-based domain parsers and homology-based domain assignment using ECOD domains found by HHsuite or Dali. Application of DPAM to the massive AlphaFold models will enable efficient classification of domains, providing evolutionary contexts and facilitating functional studies.
Keywords: domain classification; domain parser; protein domains; structural predictions.
© 2022 The Protein Society.
Figures



References
-
- Alexandrov N, Shindyalov I. PDP: protein domain parser. Bioinformatics. 2003;19(3):429–30. - PubMed

