Graph algorithms for predicting subcellular localization at the pathway level
- PMID: 36540972
- PMCID: PMC9817068
- DOI: 10.1142/9789811270611_0014
Graph algorithms for predicting subcellular localization at the pathway level
Abstract
Protein subcellular localization is an important factor in normal cellular processes and disease. While many protein localization resources treat it as static, protein localization is dynamic and heavily influenced by biological context. Biological pathways are graphs that represent a specific biological context and can be inferred from large-scale data. We develop graph algorithms to predict the localization of all interactions in a biological pathway as an edge-labeling task. We compare a variety of models including graph neural networks, probabilistic graphical models, and discriminative classifiers for predicting localization annotations from curated pathway databases. We also perform a case study where we construct biological pathways and predict localizations of human fibroblasts undergoing viral infection. Pathway localization prediction is a promising approach for integrating publicly available localization data into the analysis of large-scale biological data.
Keywords: Probabilistic graphical model; graph neural network; spatial proteomics.
Figures

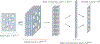
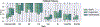

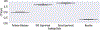
References
-
- Lundberg E and Borner GHH. Spatial proteomics: A powerful discovery tool for cell biology. Nature Reviews Molecular Cell Biology, 20(5):285–302, May 2019. - PubMed
-
- Hung M-C and Link W. Protein localization in disease and therapy. Journal of Cell Science, 124(20):3381, October 2011. - PubMed
-
- Bauer NC et al. Mechanisms regulating protein localization. Traffic, 16(10):1039–1061, 2015. - PubMed

