LdtC Is a Key l,d-Transpeptidase for Peptidoglycan Assembly in Mycobacterium smegmatis
- PMID: 36541811
- PMCID: PMC9879121
- DOI: 10.1128/jb.00424-22
LdtC Is a Key l,d-Transpeptidase for Peptidoglycan Assembly in Mycobacterium smegmatis
Abstract
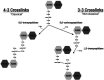
The peptidoglycan of mycobacteria has two types of direct cross-links, classical 4-3 cross-links that occur between diaminopimelate (DAP) and alanine residues, and nonclassical 3-3 cross-links that occur between DAP residues on adjacent peptides. The 3-3 cross-links are synthesized by the concerted action of d,d-carboxypeptidases and l,d-transpeptidases (Ldts). Mycobacterial genomes encode several Ldt proteins that can be classified into six classes based upon sequence identity. As a group, the Ldt enzymes are resistant to most β-lactam antibiotics but are susceptible to carbapenem antibiotics, with the exception of LdtC, a class 5 enzyme. In previous work, we showed that loss of LdtC has the greatest effect on the carbapenem susceptibility phenotype of Mycobacterium smegmatis (also known as Mycolicibacterium smegmatis) compared to other ldt deletion mutants. In this work, we show that a M. smegmatis mutant lacking the five ldt genes other than ldtC has a wild-type phenotype with the exception of increased susceptibility to rifampin. In contrast, a mutant lacking all six ldt genes has pleiotropic cell envelope defects, is temperature sensitive, and has increased susceptibility to a variety of antibiotics. These results indicate that LdtC is capable of functioning as the sole l,d-transpeptidase in M. smegmatis and suggest that it may represent a carbapenem-resistant pathway for peptidoglycan biosynthesis. IMPORTANCE Mycobacteria have several enzymes to catalyze nonclassical 3-3 linkages in the cell wall peptidoglycan. Understanding the biology of these cross-links is important for the development of antibiotic therapies to target peptidoglycan biosynthesis. Our work provides evidence that LdtC can function as the sole enzyme for 3-3 cross-link formation in M. smegmatis and suggests that LdtC may be part of a carbapenem-resistant l,d-transpeptidase pathway.
Keywords: antibiotic resistance; cell wall; l,d-transpeptidase; mycobacteria; peptidoglycan.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- World Health Organization. 2017. Global tuberculosis report.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

