Dissecting the general mechanisms of protein cage self-assembly by coarse-grained simulations
- PMID: 36541820
- PMCID: PMC9854185
- DOI: 10.1002/pro.4552
Dissecting the general mechanisms of protein cage self-assembly by coarse-grained simulations
Abstract
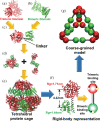
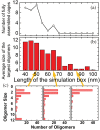
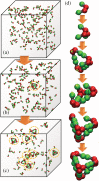
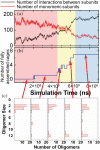
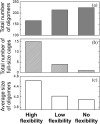
The development of artificial protein cages has recently gained massive attention due to their promising application prospect as novel delivery vehicles for therapeutics. These nanoparticles are formed through a process called self-assembly, in which individual subunits spontaneously arrange into highly ordered patterns via non-covalent but specific interactions. Therefore, the first step toward the design of novel engineered protein cages is to understand the general mechanisms of their self-assembling dynamics. Here we have developed a new computational method to tackle this problem. Our method is based on a coarse-grained model and a diffusion-reaction simulation algorithm. Using a tetrahedral cage as test model, we showed that self-assembly of protein cage requires of a seeding process in which specific configurations of kinetic intermediate states are identified. We further found that there is a critical concentration to trigger self-assembly of protein cages. This critical concentration allows that cages can only be successfully assembled under a persistently high concentration. Additionally, phase diagram of self-assembly has been constructed by systematically testing the model across a wide range of binding parameters. Finally, our simulations demonstrated the importance of protein's structural flexibility in regulating the dynamics of cage assembly. In summary, this study throws lights on the general principles underlying self-assembly of large cage-like protein complexes and thus provides insights to design new nanomaterials.
Keywords: coarse-grained simulation; complex assembly; conformational flexibility; protein cage.
© 2022 The Protein Society.
Conflict of interest statement
The authors declare no competing financial interests.
Figures

References
-
- Ander M, Beltrao P, di Ventura B, Ferkinghoff‐Borg J, Foglierini M, Kaplan A, et al. SmartCell, a framework to simulate cellular processes that combines stochastic approximation with diffusion and localisation: analysis of simple networks. Syst Biol (Stevenage). 2004;1(1):129–38. - PubMed
-
- Andrews SS, Bray D. Stochastic simulation of chemical reactions with spatial resolution and single molecule detail. Phys Biol. 2004;1(3–4):137–51. - PubMed
-
- Aragon S. A precise boundary element method for macromolecular transport properties. J Comput Chem. 2004;25(9):1191–205. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

