CRISPR-Suppressor Scanning for Systematic Discovery of Drug-Resistance Mutations
- PMID: 36541895
- PMCID: PMC10073897
- DOI: 10.1002/cpz1.614
CRISPR-Suppressor Scanning for Systematic Discovery of Drug-Resistance Mutations
Abstract
CRISPR-Cas9 genome editing technologies have enabled complex genetic manipulations in situ, including large-scale, pooled screening approaches to probe and uncover mechanistic insights across various biological processes. The RNA-programmable nature of CRISPR-Cas9 greatly empowers tiling mutagenesis approaches to elucidate molecular details of protein function, in particular the interrogation of mechanisms of resistance to small molecules, an approach termed CRISPR-suppressor scanning. In a typical CRISPR-suppressor scanning experiment, a pooled library of single-guide RNAs is designed to target across the coding sequence(s) of one or more genes, enabling the Cas9 nuclease to systematically mutate the targeted proteins and generate large numbers of diverse protein variants in situ. This cellular pool of protein variants is then challenged with drug treatment to identify mutations conferring a fitness advantage. Drug-resistance mutations identified with this approach can not only elucidate drug mechanism of action but also reveal deeper mechanistic insights into protein structure-function relationships. In this article, we outline the framework for a standard CRISPR-suppressor scanning experiment. Specifically, we provide instructions for the design and construction of a pooled sgRNA library, execution of a CRISPR-suppressor scanning screen, and basic computational analysis of the resulting data. © 2022 Wiley Periodicals LLC. Basic Protocol 1: Design and generation of a pooled sgRNA library Support Protocol 1: sgRNA library design using command-line CRISPOR Support Protocol 2: Production and titering of pooled sgRNA library lentivirus Basic Protocol 2: Execution and analysis of a CRISPR-suppressor scanning experiment.
Keywords: CRISPR; CRISPR-suppressor scanning; drug resistance; functional genomics; genetic engineering; tiling mutagenesis.
© 2022 Wiley Periodicals LLC.
Conflict of interest statement
CONFLICT OF INTEREST STATEMENT:
B.B.L. is on the scientific advisory board of H3 Biomedicine, holds a sponsored research project with H3 Biomedicine, is a scientific consultant for Imago BioSciences, and is a shareholder and member of the scientific advisory board of Light Horse Therapeutics. The remaining authors declare no competing interests.
Figures
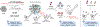

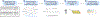


References
-
- Azam M, Latek RR, and Daley GQ 2003. Mechanisms of Autoinhibition and STI-571/Imatinib Resistance Revealed by Mutagenesis of BCR-ABL. Cell 112:831–843. - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

