Role of intraflagellar transport in transcriptional control during flagellar regeneration in Chlamydomonas
- PMID: 36542488
- PMCID: PMC10208099
- DOI: 10.1091/mbc.E22-09-0444
Role of intraflagellar transport in transcriptional control during flagellar regeneration in Chlamydomonas
Abstract
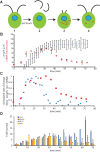
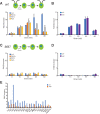
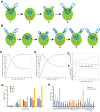
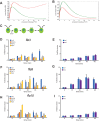
Biosynthesis of organelle precursors is a central part of the organelle size control problem, but what systems are required to control precursor production? Genes encoding flagellar proteins are up-regulated during flagellar regeneration in Chlamydomonas, and this up-regulation is critical for flagella to reach their final length, but it not known how the cell triggers these genes during regeneration. We present two models based on transcriptional repressor that is produced either in the flagellum or in the cell body and sequestered in the growing flagellum. Both models lead to stable flagellar length control and can reproduce the observed dynamics of gene expression. The two models make opposite predictions regarding the effect of mutations that block intraflagellar transport (IFT). Using quantitative measurements of gene expression, we show that gene expression during flagellar regeneration is greatly reduced in mutations of the heterotrimeric kinesin-2 that drives IFT. This result is consistent with the predictions of the model in which a repressor is sequestered in the flagellum by IFT. Inhibiting axonemal assembly has a much smaller effect on gene expression. The repressor sequestration model allows precursor production to occur when flagella are growing rapidly, representing a form of derivative control.
Figures
Similar articles
-
Testing the ion-current model for flagellar length sensing and IFT regulation.Elife. 2023 Jan 13;12:e82901. doi: 10.7554/eLife.82901. Elife. 2023. PMID: 36637158 Free PMC article.
-
Actin is required for IFT regulation in Chlamydomonas reinhardtii.Curr Biol. 2014 Sep 8;24(17):2025-32. doi: 10.1016/j.cub.2014.07.038. Epub 2014 Aug 21. Curr Biol. 2014. PMID: 25155506 Free PMC article.
-
Intraflagellar transport (IFT) cargo: IFT transports flagellar precursors to the tip and turnover products to the cell body.J Cell Biol. 2004 Jan 19;164(2):255-66. doi: 10.1083/jcb.200308132. Epub 2004 Jan 12. J Cell Biol. 2004. PMID: 14718520 Free PMC article.
-
The intraflagellar transport machinery of Chlamydomonas reinhardtii.Traffic. 2003 Jul;4(7):435-42. doi: 10.1034/j.1600-0854.2003.t01-1-00103.x. Traffic. 2003. PMID: 12795688 Review.
-
Regulation of flagellar length in Chlamydomonas.Semin Cell Dev Biol. 2008 Dec;19(6):494-501. doi: 10.1016/j.semcdb.2008.07.005. Epub 2008 Jul 25. Semin Cell Dev Biol. 2008. PMID: 18692148 Free PMC article. Review.
Cited by
-
Physical Forces in Regeneration of Cells and Tissues.Cold Spring Harb Perspect Biol. 2025 Apr 1;17(4):a041527. doi: 10.1101/cshperspect.a041527. Cold Spring Harb Perspect Biol. 2025. PMID: 38806241 Free PMC article. Review.
-
Chlamydomonas as a model system to study cilia and flagella using genetics, biochemistry, and microscopy.Front Cell Dev Biol. 2024 May 30;12:1412641. doi: 10.3389/fcell.2024.1412641. eCollection 2024. Front Cell Dev Biol. 2024. PMID: 38872931 Free PMC article. Review.
-
The flagellar length control system: exploring the physical biology of organelle size.Phys Biol. 2023 Jan 24;20(2):10.1088/1478-3975/acb18d. doi: 10.1088/1478-3975/acb18d. Phys Biol. 2023. PMID: 36623317 Free PMC article. Review.
-
Cilia and transcription: a mini review.Front Cell Dev Biol. 2025 Jun 9;13:1582796. doi: 10.3389/fcell.2025.1582796. eCollection 2025. Front Cell Dev Biol. 2025. PMID: 40552307 Free PMC article. Review.

