GABA signaling triggered by TMC-1/Tmc delays neuronal aging by inhibiting the PKC pathway in C. elegans
- PMID: 36542715
- PMCID: PMC9770988
- DOI: 10.1126/sciadv.adc9236
GABA signaling triggered by TMC-1/Tmc delays neuronal aging by inhibiting the PKC pathway in C. elegans
Abstract
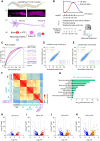
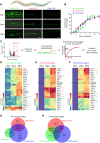
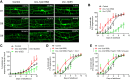
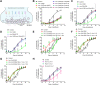
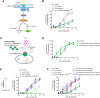
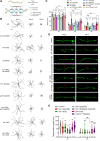
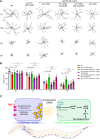
Aging causes functional decline and degeneration of neurons and is a major risk factor of neurodegenerative diseases. To investigate the molecular mechanisms underlying neuronal aging, we developed a new pipeline for neuronal proteomic profiling in young and aged animals. While the overall translational machinery is down-regulated, certain proteins increase expressions upon aging. Among these aging-up-regulated proteins, the conserved channel protein TMC-1/Tmc has an anti-aging function in all neurons tested, and the neuroprotective function of TMC-1 occurs by regulating GABA signaling. Moreover, our results show that metabotropic GABA receptors and G protein GOA-1/Goα are required for the anti-neuronal aging functions of TMC-1 and GABA, and the activation of GABA receptors prevents neuronal aging by inhibiting the PLCβ-PKC pathway. Last, we show that the TMC-1-GABA-PKC signaling axis suppresses neuronal functional decline caused by a pathogenic form of human Tau protein. Together, our findings reveal the neuroprotective function of the TMC-1-GABA-PKC signaling axis in aging and disease conditions.
Figures



References
-
- J. V. Pluvinage, T. Wyss-Coray, Systemic factors as mediators of brain homeostasis, ageing and neurodegeneration. Nat. Rev. Neurosci. 21, 93–102 (2020). - PubMed
-
- Y. Hou, X. Dan, M. Babbar, Y. Wei, S. G. Hasselbalch, D. L. Croteau, V. A. Bohr, Ageing as a risk factor for neurodegenerative disease. Nat. Rev. Neurol. 15, 565–581 (2019). - PubMed
-
- Ageing - United Nations report on world population ageing. World Today 58, 12–13 (2002).
-
- F. Sierra, E. Hadley, R. Suzman, R. Hodes, Prospects for life span extension. Annu. Rev. Med. 60, 457–469 (2009). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

