Circulating Tumor DNA Profiling for Detection, Risk Stratification, and Classification of Brain Lymphomas
- PMID: 36542815
- PMCID: PMC10419411
- DOI: 10.1200/JCO.22.00826
Circulating Tumor DNA Profiling for Detection, Risk Stratification, and Classification of Brain Lymphomas
Abstract
Purpose: Clinical outcomes of patients with CNS lymphomas (CNSLs) are remarkably heterogeneous, yet identification of patients at high risk for treatment failure is challenging. Furthermore, CNSL diagnosis often remains unconfirmed because of contraindications for invasive stereotactic biopsies. Therefore, improved biomarkers are needed to better stratify patients into risk groups, predict treatment response, and noninvasively identify CNSL.
Patients and methods: We explored the value of circulating tumor DNA (ctDNA) for early outcome prediction, measurable residual disease monitoring, and surgery-free CNSL identification by applying ultrasensitive targeted next-generation sequencing to a total of 306 tumor, plasma, and CSF specimens from 136 patients with brain cancers, including 92 patients with CNSL.
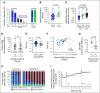
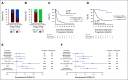
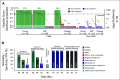
Results: Before therapy, ctDNA was detectable in 78% of plasma and 100% of CSF samples. Patients with positive ctDNA in pretreatment plasma had significantly shorter progression-free survival (PFS, P < .0001, log-rank test) and overall survival (OS, P = .0001, log-rank test). In multivariate analyses including established clinical and radiographic risk factors, pretreatment plasma ctDNA concentrations were independently prognostic of clinical outcomes (PFS HR, 1.4; 95% CI, 1.0 to 1.9; P = .03; OS HR, 1.6; 95% CI, 1.1 to 2.2; P = .006). Moreover, measurable residual disease detection by plasma ctDNA monitoring during treatment identified patients with particularly poor prognosis following curative-intent immunochemotherapy (PFS, P = .0002; OS, P = .004, log-rank test). Finally, we developed a proof-of-principle machine learning approach for biopsy-free CNSL identification from ctDNA, showing sensitivities of 59% (CSF) and 25% (plasma) with high positive predictive value.
Conclusion: We demonstrate robust and ultrasensitive detection of ctDNA at various disease milestones in CNSL. Our findings highlight the role of ctDNA as a noninvasive biomarker and its potential value for personalized risk stratification and treatment guidance in patients with CNSL.
[Media: see text].
Conflict of interest statement
The following represents disclosure information provided by authors of this manuscript. All relationships are considered compensated unless otherwise noted. Relationships are self-held unless noted. I = Immediate Family Member, Inst = My Institution. Relationships may not relate to the subject matter of this manuscript. For more information about ASCO's conflict of interest policy, please refer to
Open Payments is a public database containing information reported by companies about payments made to US-licensed physicians (
No other potential conflicts of interest were reported.
Figures

Comment in
-
Circulating Tumor DNA in the Blood: A New Frontier in Primary CNS Lymphoma?J Clin Oncol. 2023 Mar 20;41(9):1649-1651. doi: 10.1200/JCO.22.02605. Epub 2023 Jan 20. J Clin Oncol. 2023. PMID: 36669147 Free PMC article. No abstract available.
References
-
- Abrey LE, Ben-Porat L, Panageas KS, et al. : Primary central nervous system lymphoma: The Memorial Sloan-Kettering Cancer Center prognostic model. J Clin Oncol 24:5711-5715, 2006 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

