Spatial heterogeneity of infiltrating T cells in high-grade serous ovarian cancer revealed by multi-omics analysis
- PMID: 36543113
- PMCID: PMC9798026
- DOI: 10.1016/j.xcrm.2022.100856
Spatial heterogeneity of infiltrating T cells in high-grade serous ovarian cancer revealed by multi-omics analysis
Abstract
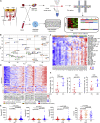
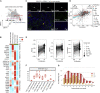
Tumor-infiltrating lymphocytes (TILs), especially CD8+ TILs, represent a favorable prognostic factor in high-grade serous ovarian cancer (HGSOC) and other tumor lineages. Here, we analyze the spatial heterogeneity of different TIL subtypes in HGSOC. We integrated RNA sequencing, whole-genome sequencing, bulk T cell receptor (TCR) sequencing, as well as single-cell RNA/TCR sequencing to investigate the characteristics and differential composition of TILs across different HGSOC sites. Two immune "cold" patterns in ovarian cancer are identified: (1) ovarian lesions with low infiltration of mainly dysfunctional T cells and immunosuppressive Treg cells and (2) omental lesions infiltrated with non-tumor-specific bystander cells. Exhausted CD8 T cells that are preferentially enriched in ovarian tumors exhibit evidence for expansion and cytotoxic activity. Inherent tumor immune microenvironment characteristics appear to be the main contributor to the spatial differences in TIL status. The landscape of spatial heterogeneity of TILs may inform potential strategies for therapeutic manipulation in HGSOC.
Keywords: TILs; high-grade serous ovarian cancer; multi-omics; multiple lesions; spatial heterogeneity.
Copyright © 2022 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests G.B.M. has licensed an HRD assay to Myriad Genetics and on Digital Spatial Profiling to Nanostring and is an SAB member/consultant with Amphista, AstraZeneca, Chrysallis Biotechnology, GSK, ImmunoMET, Ionis, Lilly, PDX Pharmaceuticals, Signalchem Lifesciences, Symphogen, Tarveda, Turbine, and Zentalis Pharmaceuticals.
Figures






References
-
- Ferlay J., Soerjomataram I., Dikshit R., Eser S., Mathers C., Rebelo M., Parkin D.M., Forman D., Bray F. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int. J. Cancer. 2015;136:E359–E386. - PubMed
-
- Sharma P., Allison J.P. The future of immune checkpoint therapy. Science. 2015;348:56–61. - PubMed
-
- González-Martín A., Sánchez-Lorenzo L. Immunotherapy with checkpoint inhibitors in patients with ovarian cancer: still promising? Cancer. 2019;125:4616–4622. - PubMed
-
- Jayson G.C., Kohn E.C., Kitchener H.C., Ledermann J.A. Ovarian cancer. Lancet. 2014;384:1376–1388. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials

