Targeting LPS biosynthesis and transport in gram-negative bacteria in the era of multi-drug resistance
- PMID: 36543281
- PMCID: PMC9922520
- DOI: 10.1016/j.bbamcr.2022.119407
Targeting LPS biosynthesis and transport in gram-negative bacteria in the era of multi-drug resistance
Abstract
Gram-negative bacteria pose a major threat to human health in an era fraught with multi-drug resistant bacterial infections. Despite extensive drug discovery campaigns over the past decades, no new antibiotic target class effective against gram-negative bacteria has become available to patients since the advent of the carbapenems in 1985. Antibiotic discovery efforts against gram-negative bacteria have been hampered by limited intracellular accumulation of xenobiotics, in large part due to the impermeable cell envelope comprising lipopolysaccharide (LPS) in the outer leaflet of the outer membrane, as well as a panoply of efflux pumps. The biosynthesis and transport of LPS are essential to the viability and virulence of most gram-negative bacteria. Thus, both LPS biosynthesis and transport are attractive pathways to target therapeutically. In this review, we summarize the LPS biosynthesis and transport pathways and discuss efforts to find small molecule inhibitors against targets within these pathways.
Keywords: Antibiotics; Cell envelope; Drug resistance; Gram-negative; Lipopolysaccharide.
Copyright © 2022 The Authors. Published by Elsevier B.V. All rights reserved.
Conflict of interest statement
Declaration of competing interest The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures

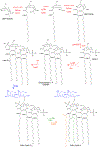



References
-
- Breidenstein EB, de la Fuente-Nunez C, Hancock RE, Pseudomonas aeruginosa: all roads lead to resistance, Trends Microbiol. 19 (2011) 419–426. - PubMed
-
- Willyard C, The drug-resistant bacteria that pose the greatest health threats, Nature 543 (2017) 15. - PubMed
-
- Tacconelli E, Carrara E, Savoldi A, Harbarth S, Mendelson M, Monnet DL, Pulcini C, Kahlmeter G, Kluytmans J, Carmeli Y, et al., Discovery, research, and development of new antibiotics: the WHO priority list of antibiotic-resistant bacteria and tuberculosis, Lancet Infect. Dis 18 (2018) 318–327. - PubMed
-
- Payne DJ, Gwynn MN, Holmes DJ, Pompliano DL, Drugs for bad bugs: confronting the challenges of antibacterial discovery, Nat. Rev. Drug Discov 6 (2007) 29–40. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

