Reconstruction and deconstruction of human somitogenesis in vitro
- PMID: 36543321
- PMCID: PMC10018515
- DOI: 10.1038/s41586-022-05655-4
Reconstruction and deconstruction of human somitogenesis in vitro
Abstract
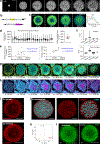
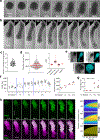
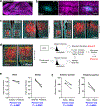
The vertebrate body displays a segmental organization that is most conspicuous in the periodic organization of the vertebral column and peripheral nerves. This metameric organization is first implemented when somites, which contain the precursors of skeletal muscles and vertebrae, are rhythmically generated from the presomitic mesoderm. Somites then become subdivided into anterior and posterior compartments that are essential for vertebral formation and segmental patterning of the peripheral nervous system1-4. How this key somitic subdivision is established remains poorly understood. Here we introduce three-dimensional culture systems of human pluripotent stem cells called somitoids and segmentoids, which recapitulate the formation of somite-like structures with anteroposterior identity. We identify a key function of the segmentation clock in converting temporal rhythmicity into the spatial regularity of anterior and posterior somitic compartments. We show that an initial 'salt and pepper' expression of the segmentation gene MESP2 in the newly formed segment is transformed into compartments of anterior and posterior identity through an active cell-sorting mechanism. Our research demonstrates that the major patterning modules that are involved in somitogenesis, including the clock and wavefront, anteroposterior polarity patterning and somite epithelialization, can be dissociated and operate independently in our in vitro systems. Together, we define a framework for the symmetry-breaking process that initiates somite polarity patterning. Our work provides a platform for decoding general principles of somitogenesis and advancing knowledge of human development.
© 2022. The Author(s), under exclusive licence to Springer Nature Limited.
Conflict of interest statement
Competing interests
The authors declare the following competing interests: O.P. is scientific founder of Anagenesis Biotechnologies. All other authors declare no competing interests.
Figures











Comment in
-
Human stem cell models unravel mechanisms of somite segmentation.Cell Stem Cell. 2023 Mar 2;30(3):246-247. doi: 10.1016/j.stem.2023.01.011. Cell Stem Cell. 2023. PMID: 36868192
References
-
- Hubaud A & Pourquié O Signalling dynamics in vertebrate segmentation. Nat. Rev. Mol. Cell Biol 15, 709–721 (2014). - PubMed
-
- Saga Y The mechanism of somite formation in mice. Curr. Opin. Genet. Dev 22, 331–338 (2012). - PubMed
-
- Fleming A, Kishida MG, Kimmel CB & Keynes RJ Building the backbone: the development and evolution of vertebral patterning . Development 142, 1733–1744 (2015). - PubMed
-
- Kuan C-YK, Tannahill D, Cook GMW & Keynes RJ Somite polarity and segmental patterning of the peripheral nervous system. Mech. Dev 121, 1055–1068 (2004). - PubMed
-
- Oates AC, Morelli LG & Ares S Patterning embryos with oscillations: structure, function and dynamics of the vertebrate segmentation clock. Development 139, 625–639 (2012). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

