A spatially resolved atlas of the human lung characterizes a gland-associated immune niche
- PMID: 36543915
- PMCID: PMC9839452
- DOI: 10.1038/s41588-022-01243-4
A spatially resolved atlas of the human lung characterizes a gland-associated immune niche
Abstract
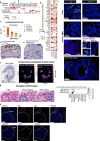
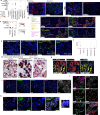
Single-cell transcriptomics has allowed unprecedented resolution of cell types/states in the human lung, but their spatial context is less well defined. To (re)define tissue architecture of lung and airways, we profiled five proximal-to-distal locations of healthy human lungs in depth using multi-omic single cell/nuclei and spatial transcriptomics (queryable at lungcellatlas.org ). Using computational data integration and analysis, we extend beyond the suspension cell paradigm and discover macro and micro-anatomical tissue compartments including previously unannotated cell types in the epithelial, vascular, stromal and nerve bundle micro-environments. We identify and implicate peribronchial fibroblasts in lung disease. Importantly, we discover and validate a survival niche for IgA plasma cells in the airway submucosal glands (SMG). We show that gland epithelial cells recruit B cells and IgA plasma cells, and promote longevity and antibody secretion locally through expression of CCL28, APRIL and IL-6. This new 'gland-associated immune niche' has implications for respiratory health.
© 2022. The Author(s).
Conflict of interest statement
In the past three years, SAT has received remuneration for consulting and Scientific Advisory Board Membership from Genentech, Roche, Biogen, GlaxoSmithKline, Foresite Labs and Qiagen. SAT is a cofounder, board member and holds equity in Transition Bio. OS is a paid member of the Scientific Advisory Board of Insitro. The remaining authors declare no competing interests.
Figures














Comment in
-
Spatial atlas reveals insight into lung immunity.Nat Genet. 2023 Jan;55(1):10-11. doi: 10.1038/s41588-022-01244-3. Nat Genet. 2023. PMID: 36609699 No abstract available.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

