Identification of novel candidate genes and predicted miRNAs in atopic dermatitis patients by bioinformatic methods
- PMID: 36543921
- PMCID: PMC9772328
- DOI: 10.1038/s41598-022-26689-8
Identification of novel candidate genes and predicted miRNAs in atopic dermatitis patients by bioinformatic methods
Abstract
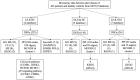
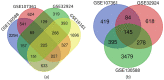
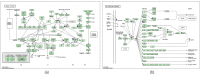
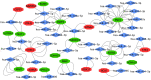
Atopic dermatitis (AD) is a common, chronic inflammatory dermatosis with relapsing eruptions. Our study used bioinformatics to find novel candidate differentially expressed genes (DEGs) and predicted miRNAs between AD patients and healthy controls. The Mesh term "atopic dermatitis" was retrieved to obtain DEGs in GEO datasets. DEGs between AD patients and healthy controls were analyzed using GEO2R. Overlapping DEGs between different datasets were obtained with use of Draw Venn software. GO and KEGG enrichment analyses were conducted by the use of DAVID. STRING and miRWalk were used to individually analyze PPI networks, interactions of candidate genes and predicted miRNAs. A total of 571 skin samples, as retrieved from 9 databases were assessed. There were 225 overlapping DEGs between lesioned skin samples of AD patients and that of healthy controls. Nineteen nodes and 160 edges were found in the largest PPI cluster, consisting of 17 up-regulated and 2 down-regulated nodes. Two KEGG pathways were identified, including the cell cycle (CCNB1, CHEK1, BUB1B, MCM5) and p53 (CCNB1, CHEK1, GTSE1) pathways. There were 56 nodes and 100 edges obtained in the miRNA-target gene network, with has-miR-17-5p targeted to 4 genes and has-miR-106b-5p targeted to 3 genes. While these findings will require further verification as achieved with experiments involving in vivo and in vitro modles, these results provided some initial insights into dysfunctional inflammatory and immune responses associated with AD. Such information offers the potential to develop novel therapeutic targets for use in preventing and treating AD.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


Similar articles
-
Identification and interaction analysis of key genes and microRNAs in atopic dermatitis by bioinformatics analysis.Clin Exp Dermatol. 2019 Apr;44(3):257-264. doi: 10.1111/ced.13691. Epub 2018 Jul 3. Clin Exp Dermatol. 2019. PMID: 29974487
-
Key Genes Associated with Pyroptosis in Gout and Construction of a miRNA-mRNA Regulatory Network.Cells. 2022 Oct 17;11(20):3269. doi: 10.3390/cells11203269. Cells. 2022. PMID: 36291136 Free PMC article.
-
Screening differentially expressed genes and the pathogenesis in atopic dermatitis using bioinformatics.Cell Mol Biol (Noisy-le-grand). 2023 Dec 31;69(15):73-78. doi: 10.14715/cmb/2023.69.15.12. Cell Mol Biol (Noisy-le-grand). 2023. PMID: 38279492
-
Construction of the miRNA-mRNA regulatory network and analysis of hub genes in oral squamous cell carcinoma.Biomed Pap Med Fac Univ Palacky Olomouc Czech Repub. 2022 Sep;166(3):280-289. doi: 10.5507/bp.2022.001. Epub 2022 Feb 3. Biomed Pap Med Fac Univ Palacky Olomouc Czech Repub. 2022. PMID: 35132271
-
Identification and interaction analysis of key genes and microRNAs in hepatocellular carcinoma by bioinformatics analysis.World J Surg Oncol. 2017 Mar 16;15(1):63. doi: 10.1186/s12957-017-1127-2. World J Surg Oncol. 2017. PMID: 28302149 Free PMC article.
Cited by
-
Co-Expression Network and Machine Learning Analysis of Transcriptomics Data Identifies Distinct Gene Signatures and Pathways in Lesional and Non-Lesional Atopic Dermatitis.J Pers Med. 2024 Sep 10;14(9):960. doi: 10.3390/jpm14090960. J Pers Med. 2024. PMID: 39338214 Free PMC article.
References
-
- Willems A, Tapley A, Fielding A, Tng ETV, Holliday EG, van Driel ML, et al. Prevalence and associations of general practice registrars’ management of atopic dermatitis: A cross-sectional analysis from the registrar clinical encounters in training study. Dermatol Pract Concept. 2021;11:e2021128. doi: 10.5826/dpc.1104a128. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

