Hypoxia drives CD39-dependent suppressor function in exhausted T cells to limit antitumor immunity
- PMID: 36543958
- PMCID: PMC10402660
- DOI: 10.1038/s41590-022-01379-9
Hypoxia drives CD39-dependent suppressor function in exhausted T cells to limit antitumor immunity
Abstract
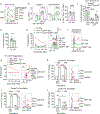
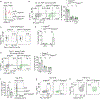
CD8+ T cells are critical for elimination of cancer cells. Factors within the tumor microenvironment (TME) can drive these cells to a hypofunctional state known as exhaustion. The most terminally exhausted T (tTex) cells are resistant to checkpoint blockade immunotherapy and might instead limit immunotherapeutic efficacy. Here we show that intratumoral CD8+ tTex cells possess transcriptional features of CD4+Foxp3+ regulatory T cells and are similarly capable of directly suppressing T cell proliferation ex vivo. tTex cell suppression requires CD39, which generates immunosuppressive adenosine. Restricted deletion of CD39 in endogenous CD8+ T cells resulted in slowed tumor progression, improved immunotherapy responsiveness and enhanced infiltration of transferred tumor-specific T cells. CD39 is induced on tTex cells by tumor hypoxia, thus mitigation of hypoxia limits tTex suppression. Together, these data suggest tTex cells are an important regulatory population in cancer and strategies to limit their generation, reprogram their immunosuppressive state or remove them from the TME might potentiate immunotherapy.
© 2022. The Author(s), under exclusive licence to Springer Nature America, Inc.
Conflict of interest statement
COMPETING INTERESTS STATEMENT
A.V.M. is currently an employee of Novasenta. G.M.D. declares competing financial interests and has submitted patents targeting exhausted T cells that are licensed or pending and is entitled to a share in net income generated from licensing of these patent rights for commercial development. G.M.D. consults for and/or is on the scientific advisory board of BlueSphere Bio, Century Therapeutics, Nanna Therapeutics, Novasenta, Pieris Pharmaceuticals, and Western Oncolytics/Kalivir; has grants from bluebird bio, Novasenta, Pfizer, Pieris Pharmaceuticals, TCR2, and Western Oncolytics/Kalivir; G.M.D. owns stock in Novasenta, BlueSphere Bio, and RemplirBio. The remaining authors have no competing interests.
Figures


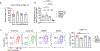






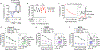






Comment in
-
Exhausted T cells in tumors gain suppressor activity and can restrain immunity.Nat Immunol. 2023 Feb;24(2):218-219. doi: 10.1038/s41590-022-01409-6. Nat Immunol. 2023. PMID: 36650281 No abstract available.
References
-
- Bruni D, Angell HK & Galon J The immune contexture and Immunoscore in cancer prognosis and therapeutic efficacy. Nat. Rev. Cancer 20, 662–680 (2020). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- DP2 AI136598/AI/NIAID NIH HHS/United States
- F31 AI149971/AI/NIAID NIH HHS/United States
- T32 CA082084/CA/NCI NIH HHS/United States
- P30 CA047904/CA/NCI NIH HHS/United States
- R01 AI171483/AI/NIAID NIH HHS/United States
- K00 CA222711/CA/NCI NIH HHS/United States
- T32 GM008208/GM/NIGMS NIH HHS/United States
- F99 CA222711/CA/NCI NIH HHS/United States
- R01 CA277473/CA/NCI NIH HHS/United States
- P50 CA097190/CA/NCI NIH HHS/United States
- R01 AI166598/AI/NIAID NIH HHS/United States
- F30 CA247034/CA/NCI NIH HHS/United States
- P50 CA121973/CA/NCI NIH HHS/United States
- F31 CA247129/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical
Research Materials

