A survey on clinical natural language processing in the United Kingdom from 2007 to 2022
- PMID: 36544046
- PMCID: PMC9770568
- DOI: 10.1038/s41746-022-00730-6
A survey on clinical natural language processing in the United Kingdom from 2007 to 2022
Abstract
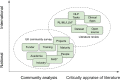
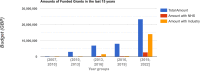
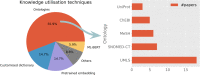
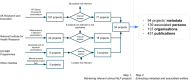
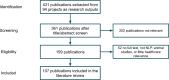
Much of the knowledge and information needed for enabling high-quality clinical research is stored in free-text format. Natural language processing (NLP) has been used to extract information from these sources at scale for several decades. This paper aims to present a comprehensive review of clinical NLP for the past 15 years in the UK to identify the community, depict its evolution, analyse methodologies and applications, and identify the main barriers. We collect a dataset of clinical NLP projects (n = 94; £ = 41.97 m) funded by UK funders or the European Union's funding programmes. Additionally, we extract details on 9 funders, 137 organisations, 139 persons and 431 research papers. Networks are created from timestamped data interlinking all entities, and network analysis is subsequently applied to generate insights. 431 publications are identified as part of a literature review, of which 107 are eligible for final analysis. Results show, not surprisingly, clinical NLP in the UK has increased substantially in the last 15 years: the total budget in the period of 2019-2022 was 80 times that of 2007-2010. However, the effort is required to deepen areas such as disease (sub-)phenotyping and broaden application domains. There is also a need to improve links between academia and industry and enable deployments in real-world settings for the realisation of clinical NLP's great potential in care delivery. The major barriers include research and development access to hospital data, lack of capable computational resources in the right places, the scarcity of labelled data and barriers to sharing of pretrained models.
© 2022. The Author(s).
Conflict of interest statement
R.S. declares research support received in the last 3 years, from Janssen, GSK and Takeda. The remaining authors declare no competing interests.
Figures





Comment in
-
Natural language processing - relevance to patient outcomes and real-world evidence.Expert Rev Pharmacoecon Outcomes Res. 2024 Jan;24(1):5-9. doi: 10.1080/14737167.2023.2275670. Epub 2024 Jan 18. Expert Rev Pharmacoecon Outcomes Res. 2024. PMID: 37874661 No abstract available.
References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

