Diversity of bacterial communities in the plasmodia of myxomycetes
- PMID: 36544088
- PMCID: PMC9773492
- DOI: 10.1186/s12866-022-02725-5
Diversity of bacterial communities in the plasmodia of myxomycetes
Abstract
Background: Myxomycetes are a group of eukaryotes belonging to Amoebozoa, which are characterized by a distinctive life cycle, including the plasmodium stage and fruit body stage. Plasmodia are all found to be associated with bacteria. However, the information about bacteria diversity and composition in different plasmodia was limited. Therefore, this study aimed to investigate the bacterial diversity of plasmodia from different myxomycetes species and reveal the potential function of plasmodia-associated bacterial communities.
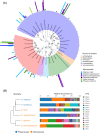
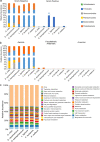
Results: The bacterial communities associated with the plasmodia of six myxomycetes (Didymium iridis, Didymium squamulosum, Diderma hemisphaericum, Lepidoderma tigrinum, Fuligo leviderma, and Physarum melleum) were identified by 16S rRNA amplicon sequencing. The six plasmodia harbored 38 to 52 bacterial operational taxonomic units (OTUs) that belonged to 7 phyla, 16 classes, 23 orders, 40 families, and 53 genera. The dominant phyla were Bacteroidetes, Firmicutes, and Proteobacteria. Most OTUs were shared among the six myxomycetes, while unique bacteria in each species only accounted for a tiny proportion of the total OTUs.
Conclusions: Although each of the six myxomycetes plasmodia had different bacterial community compositions, a high similarity was observed in the plasmodia-associated bacterial communities' functional composition. The high enrichment for gram-negative (> 90%) and aerobic (> 99%) bacteria in plasmodia suggest that myxomycetes may positively recruit certain kinds of bacteria from the surrounding environment.
Keywords: 16S rRNA; Amplicon sequencing; Bacteria; Slime mold.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
Metagenomic Insight into the Associated Microbiome in Plasmodia of Myxomycetes.Microorganisms. 2024 Dec 10;12(12):2540. doi: 10.3390/microorganisms12122540. Microorganisms. 2024. PMID: 39770743 Free PMC article.
-
Isolation of bioactive natural products from myxomycetes.Med Chem. 2005 Nov;1(6):575-90. doi: 10.2174/157340605774598135. Med Chem. 2005. PMID: 16787341 Review.
-
Identification of the telomeric sequence of the acellular slime molds Didymium iridis and Physarum polycephalum.Nucleic Acids Res. 1987 Nov 25;15(22):9143-52. doi: 10.1093/nar/15.22.9143. Nucleic Acids Res. 1987. PMID: 3684591 Free PMC article.
-
Multigene phylogeny of the order Physarales (Myxomycetes, Amoebozoa): shedding light on the dark-spored clade.Persoonia. 2023 Jun;51:89-124. doi: 10.3767/persoonia.2023.51.02. Epub 2023 Aug 23. Persoonia. 2023. PMID: 38665983 Free PMC article.
-
The Systematics and Phylogeny of Myxomycetes: Yesterday, Today, and Tomorrow.Dokl Biol Sci. 2024 Dec;519(1):356-369. doi: 10.1134/S0012496624701242. Epub 2024 Oct 11. Dokl Biol Sci. 2024. PMID: 39400900 Review.
Cited by
-
Slime molds (Myxomycetes) causing a "disease" in crop plants and cultivated mushrooms.Front Plant Sci. 2024 Jun 10;15:1411231. doi: 10.3389/fpls.2024.1411231. eCollection 2024. Front Plant Sci. 2024. PMID: 38916031 Free PMC article. Review.
-
Metagenomic Insight into the Associated Microbiome in Plasmodia of Myxomycetes.Microorganisms. 2024 Dec 10;12(12):2540. doi: 10.3390/microorganisms12122540. Microorganisms. 2024. PMID: 39770743 Free PMC article.
-
Spatiotemporal patterns of soil myxomycetes in subtropical managed forests and their potential interactions with bacteria.Appl Environ Microbiol. 2025 Jun 18;91(6):e0047925. doi: 10.1128/aem.00479-25. Epub 2025 May 13. Appl Environ Microbiol. 2025. PMID: 40358238 Free PMC article.
-
Didymium arenosum, a myxomycete new to science from the confluence of deserts in northwestern China.PeerJ. 2024 Jan 8;12:e16725. doi: 10.7717/peerj.16725. eCollection 2024. PeerJ. 2024. PMID: 38213774 Free PMC article.
References
-
- Stephenson SL, Schnittler M, Novozhilov YK. Myxomycete diversity and distribution from the fossil record to the present. Biodivers Conserv. 2008;17:285–301. doi: 10.1007/s10531-007-9252-9. - DOI
-
- Gilbert FA. Feeding habits of the swarm cells of the myxomycete Dictydiaethalium plumbeum. Am J Bot. 1928;15:123–131. doi: 10.1002/j.1537-2197.1928.tb04886.x. - DOI
-
- Feest A, Madelin MF. A method for the enumeration of myxomycetes in soils and its application to a wide range of soils. FEMS Microbiol Lett. 1985;31:103–109. doi: 10.1111/j.1574-6968.1985.tb01137.x. - DOI
-
- Foissner W. Soil protozoa as bioindicators: pros and cons, methods, diversity, representative examples. Agr Ecosyst Environ. 1999;74:95–112. doi: 10.1016/S0167-8809(99)00032-8. - DOI
-
- Stephenson SL. From morphological to molecular: studies of myxomycetes since the publication of the Mar-tin and Alexopoulos (1969) monograph. Fungal Divers. 2011;50:21–34. doi: 10.1007/s13225-011-0113-1. - DOI
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

